Illumina Paired End Sequencing
Illumina gets sequence data from both strands of input sequence which means it outputs data from both ends of the input and is normally reported two files R1 and R2, often refereed to as mates files (R1=first mates, R2=second mates). Due to the way data is reported in these files, special care has to be taken when processing these data files. What ends up in the first mates files vs the second mates files is dependent on how the forward and reverse Illumina adapter/primers are added to the sequences with sequence immediately following the forward Illumina adapter/primer being in R1 and the sequence that comes right before the reverse Illumina adapter/primer being in R2. This adapter sequence is most commonly trimmed off by Illumina’s own software associated with the sequencing machine itself.

Overlap status
Depending on the length of your input and the length being sequences there are several ways your sequences in the mate files end up overlapping if there is any overlapping at all.
- no overlap the sequences do not overlap
- first mate ends in the second mate
- first mate begins in the second mate (in this case the primer/adapters from the other ends are sequences, a scenario referred to as “read through”)

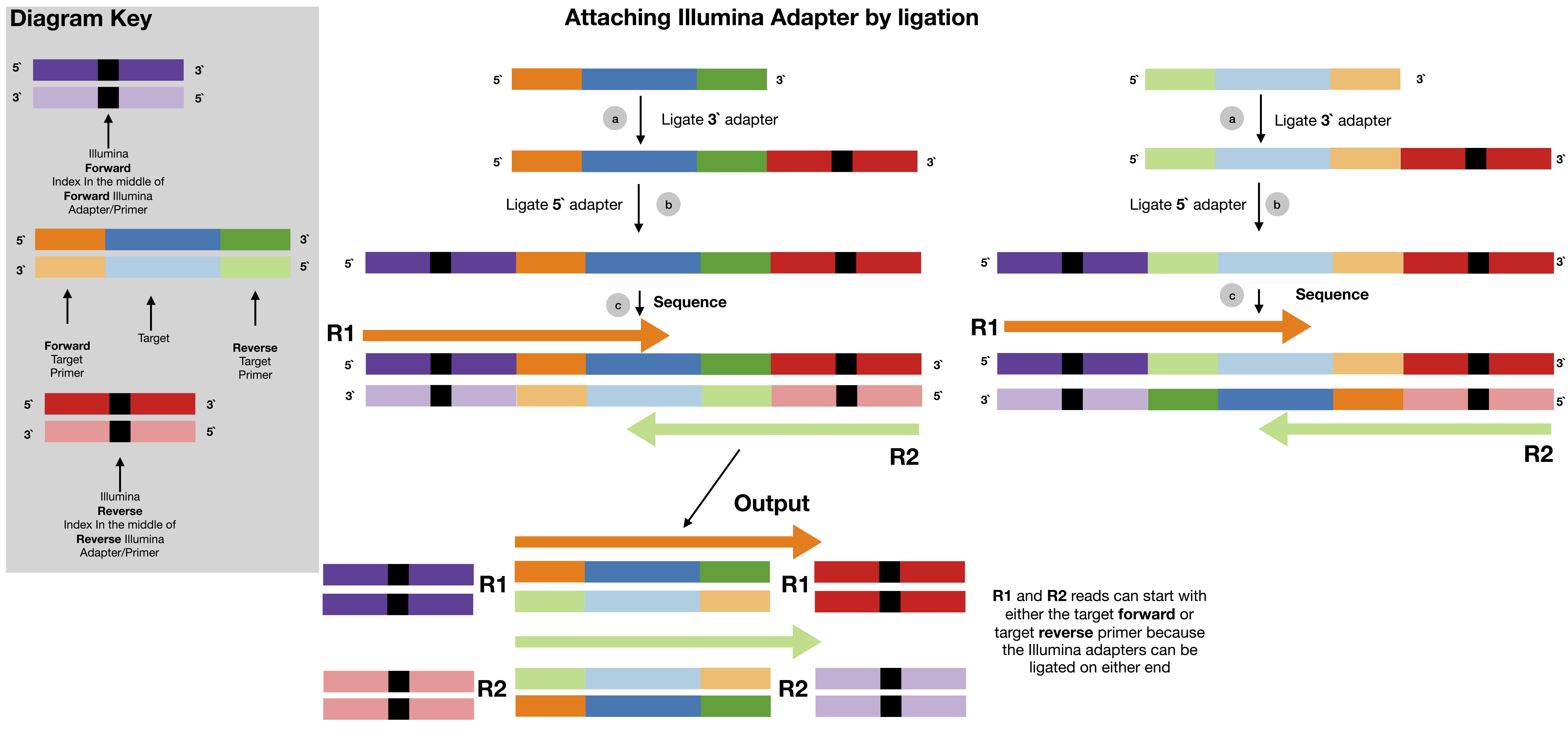
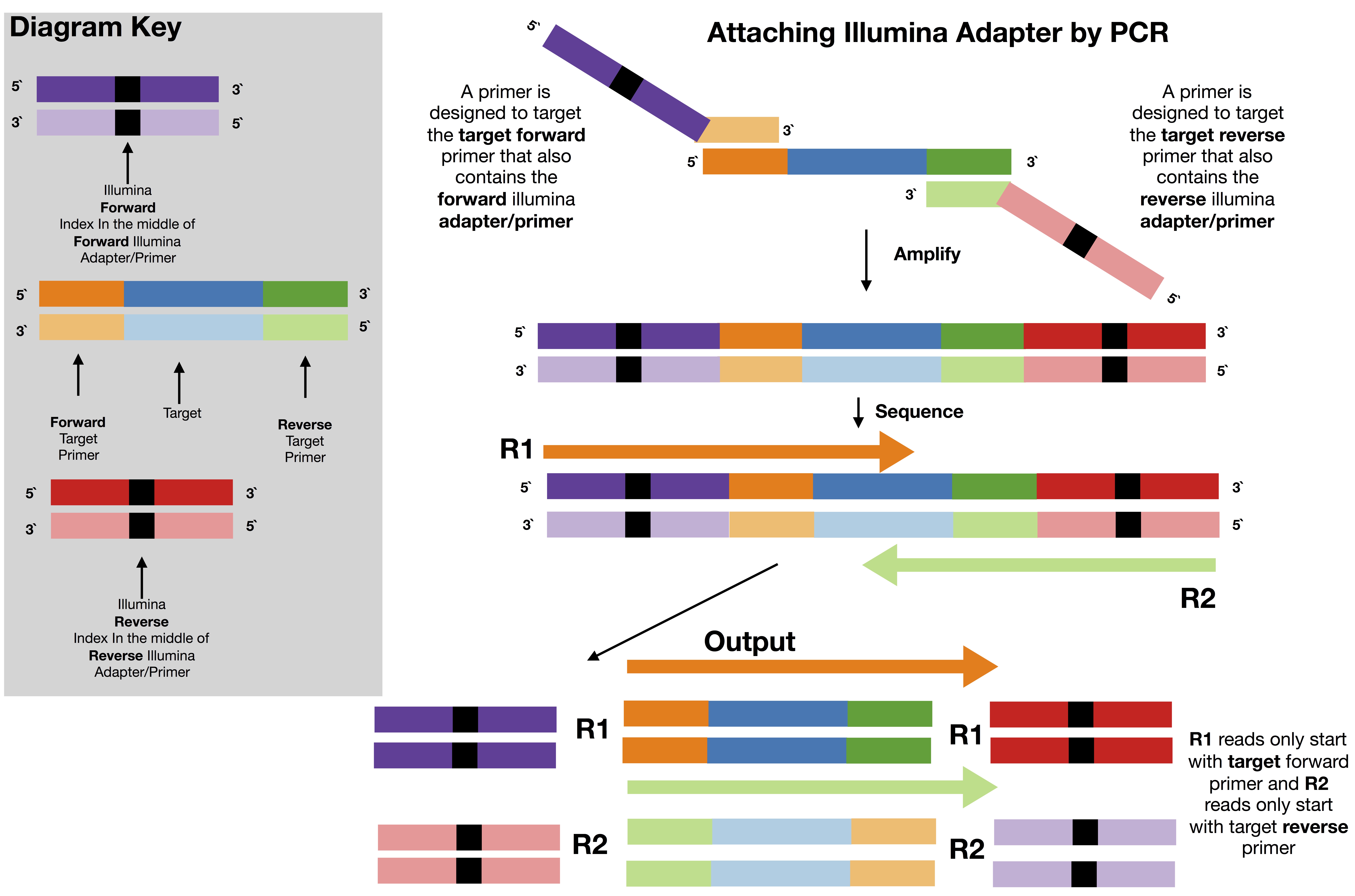
Attaching Illumina adapters/primers
Depending on how Illumina adapters/primers are attached to your target of interest can affect the orientation of the sequences you get from the machine.
By PCR
If Illumina adapters/primers are attached by PCR, depending on how this is done all of your final sequences can end up in the same “direction”.
By ligation
If Illumina adapters/primers are attached by ligation by simply attaching using sticky ends ligation (again could depend on library prep), your final sequences can end up as a mix of both directions.