Code
load("rowAnnoColors.Rdata")load("rowAnnoColors.Rdata")meta.tab.txtmetaByBioSamplereal_mccoil_COI_calls.tsvslim_allSelectedClustersInfo.tab.txt.gzmetaSelected.tab.txtallMeta_HRP2_HRP3_deletionCalls.tab.txtsubwindows_regionMeta.tab.txt
meta = readr::read_tsv("../meta/metadata/meta.tab.txt") %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
metaByBioSample = readr::read_tsv("../meta/metadata/metaByBioSample.tab.txt") %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
# coiCalls = readr::read_tsv("/Users/nick/Dropbox (Personal)/ownCloud/documents/plasmodium/falciparum/pfepipanels/Pf_Epi_Panels/data/MAD4HATTER/data/pf/COI_calls.tab.txt")
# coiCalls_poly = coiCalls %>%
# filter(COI > 1)
coiCalls = readr::read_tsv("heome1_COI_calls.tab.txt")
#coiCalls = readr::read_tsv("PfSMART_COI_calls.tab.txt")
coiCalls_poly = coiCalls %>%
filter(COI > 1)
realmccoilCoiCalls = readr::read_tsv("wgs_variants/THEREALMcCOIL/categorical_method/real_mccoil_COI_calls.tsv")
realmccoilCoiCalls_poly = realmccoilCoiCalls %>%
filter(random_median != 1 | topHE_median != 1)
previousDeletionCalls = readr::read_tsv("allMeta_HRP2_HRP3_deletionCalls.tab.txt") %>%
#filter(country %!in% c("Bangladesh", "Mauritania", "Myanmar", "The Gambia")) %>%
#filter(((grepl("SPT", sample) & possiblyChr11Deleted))) %>%
#filter(BiologicalSample %!in% coiCalls_poly$sample) %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
meta = meta %>%
left_join(previousDeletionCalls)%>%
mutate(hrpCall = case_when(
possiblyHRP2Deleted & possiblyHRP3Deleted ~ "pfhrp2-/pfhrp3-",
possiblyHRP2Deleted & !possiblyHRP3Deleted ~ "pfhrp2-/pfhrp3+",
!possiblyHRP2Deleted & possiblyHRP3Deleted ~ "pfhrp2+/pfhrp3-",
T ~ "pfhrp2+/pfhrp3+"
)) %>%
left_join(realmccoilCoiCalls %>%
select(BiologicalSample, topHE_median) %>%
rename(COI = topHE_median))
# left_join(coiCalls %>%
# rename(BiologicalSample = sample))
homologousRegion = readr::read_tsv("../rRNA_segmental_duplications/sharedBetween11_and_13/investigatingChrom11Chrom13/Pf3D7_13_v3-2792021-2807295-for--Pf3D7_11_v3-1918028-1933288-for.bed",
col_names = F)
regions = readr::read_tsv("subwindows_regionMeta.tab.txt")
metaSelected = readr::read_tsv("metaSelected.tab.txt") %>%
#select(-COI) %>%
left_join(realmccoilCoiCalls %>%
select(BiologicalSample, topHE_median) %>%
rename(COI = topHE_median)) %>%
filter(COI == 1)
metaSelected_hrp2_deleted = metaSelected %>% filter(possiblyHRP2Deleted)
metaSelected_hrp3_deleted = metaSelected %>% filter(possiblyHRP3Deleted)
metaSelected_hrp2_and_hrp3_deleted = metaSelected %>% filter(possiblyHRP2Deleted, possiblyHRP3Deleted)
regions_key = regions %>%
select(name, genomicID)finalHrpSubwindows = readr::read_tsv("../windowAnalysis/windows/finalHRPII_HRPIII_windows_withTuned_combinedVarConservedRegions.bed", col_names = F)
erroneousRegions = c("Pf3D7_11_v3-1944071-1944237", "Pf3D7_11_v3-1944083-1944229", "Pf3D7_11_v3-1938175-1938354")
samplesCovered = readr::read_tsv("samplesCovered.txt", col_names = "sample") %>%
left_join(meta %>%
select(sample, BiologicalSample))popClustering = readr::read_tsv("finalHRPII_HRPIII_windows_withTunedSubWindows/popClustering/reports/slim_allSelectedClustersInfo.tab.txt.gz")
#
regions_key = regions_key %>%
mutate(duplicationRegion = grepl("for", name))
# renaming and duplicate the dup region
popClustering = popClustering %>%
left_join(regions_key %>%
rename(p_name = name)) %>%
mutate(p_name = genomicID) %>%
mutate(h_popUID = paste0(genomicID, "--", h_popUID))
#
popClustering_filt = popClustering %>%
filter(s_Sample %fin% metaSelected$BiologicalSample) %>%
filter(genomicID %!in% erroneousRegions)
previousDeletionCalls_hrp3Delete = previousDeletionCalls %>%
filter(possiblyHRP3Deleted)
popClustering_filt_hrp3Delete = popClustering_filt %>%
filter(s_Sample %in% previousDeletionCalls_hrp3Delete$BiologicalSample)
regions_afterHomologous = regions %>%
filter(afterHomologousRegion)allDeletionTypeMeta = readr::read_tsv("allMetaDeletionCalls.tab.txt") %>%
filter(BiologicalSample %in% metaSelected$BiologicalSample)
allDeletionTypeMeta_hrp3_pat1 = allDeletionTypeMeta %>%
filter(HRP3_deletionPattern == "Pattern 1")
popClustering_filt_hrp3_pat1 = popClustering_filt %>%
filter(s_Sample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample)
allDeletionTypeMeta_hrp3_pat2 = allDeletionTypeMeta %>%
filter(HRP3_deletionPattern == "Pattern 2")
popClustering_filt_hrp3_pat2 = popClustering_filt %>%
filter(s_Sample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample)
allDeletionTypeMeta_deletionPatternCounts = allDeletionTypeMeta %>%
filter(!is.na(HRP3_deletionPattern)) %>%
group_by(HRP3_deletionPattern) %>%
count()
create_dt(allDeletionTypeMeta_deletionPatternCounts)allDeletionTypeMeta_hrp3_pat2_count_country = allDeletionTypeMeta_hrp3_pat2 %>%
group_by(country, region, secondaryRegion) %>%
count()
create_dt(allDeletionTypeMeta_hrp3_pat2_count_country)allDeletionTypeMeta_hrp3_pat2_count_region = allDeletionTypeMeta_hrp3_pat2 %>%
group_by(region, secondaryRegion) %>%
count()
create_dt(allDeletionTypeMeta_hrp3_pat2_count_region)allDeletionTypeMeta_hrp3_pat2_count_continent = allDeletionTypeMeta_hrp3_pat2 %>%
group_by(secondaryRegion) %>%
count()
create_dt(allDeletionTypeMeta_hrp3_pat2_count_continent)Below is code determining the samples with possible chr11 fragment duplication and breaking down the counts of perfect duplicated copies vs divergent copies.
regions_afterHomologous_chr11 = regions %>%
filter(`#chrom` == "Pf3D7_11_v3",
afterHomologousRegion)
regions_afterHomologous_chr11 = regions_afterHomologous_chr11 %>%
mutate(description = case_when(
grepl("extraField0=NA", extraField0) ~ "intergenic",
T ~ gsub("\\]", "", gsub(".*description=", "", extraField0))
) )
descriptionColors = scheme$hex(length(regions_afterHomologous_chr11$description %>% unique()))
names(descriptionColors) = regions_afterHomologous_chr11$description %>% unique()
descriptionColors["intergenic"] = c("#FF000000")popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 = popClustering_filt_hrp3_pat1 %>%
filter(p_name %in% regions_afterHomologous_chr11$genomicID)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
group_by(s_Sample, p_name) %>%
mutate(uniqHaps= n())
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_uniqSum = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
group_by(s_Sample) %>%
mutate(targets = length(unique(genomicID))) %>%
group_by(s_Sample, targets, uniqHaps) %>%
count() %>%
mutate(freq = n/targets)
minafCutoff = 0.15
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
filter(c_AveragedFrac > minafCutoff) %>%
group_by(s_Sample, p_name) %>%
mutate(uniqHaps= n()) %>%
mutate(marker = uniqHaps == 1) %>%
group_by(s_Sample) %>%
summarise(conserved = sum(marker),
targets = length(unique(genomicID))) %>%
mutate(conservedID = conserved/targets)
conservedCutOff = 0.99
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum %>%
filter(conservedID > conservedCutOff)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOff = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
group_by() %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOffByCountry = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
left_join(metaByBioSample %>%
rename(s_Sample = sample)) %>%
group_by(country, region) %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOffByRegion = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
left_join(metaByBioSample %>%
rename(s_Sample = sample)) %>%
group_by(region) %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOffByContinent = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
left_join(metaByBioSample %>%
rename(s_Sample = sample)) %>%
group_by(secondaryRegion) %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)The number of samples with perfect copies
create_dt(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOff)The number of samples with perfect copies broken down by country
create_dt(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOffByCountry)The number of samples with perfect copies broken down by regions
create_dt(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOffByRegion)The number of samples with perfect copies broken down by continent.
create_dt(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOffByContinent)The breakdown of level of divergence in the samples with divergent samples.
create_dt(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_meanId)Calculating the population of the haplotypes after the shared region on chr 11, the duplicated region to see if there is any population signal associated with the duplicated copy. E.g. if the copy is unique to a subset of haplotypes, if the copy is always perfect or if there is variation.
popClustering_filt_regions_afterHomologous_chr11 = popClustering %>%
filter(genomicID %!in% erroneousRegions) %>%
filter(p_name %in% regions_afterHomologous_chr11$genomicID)
popClustering_filt_regions_afterHomologous_chr11_tarCounts = popClustering_filt_regions_afterHomologous_chr11 %>%
group_by(s_Sample) %>%
summarise(tarCounts = length(unique(p_name)))
popClustering_filt_regions_afterHomologous_chr11_tarCounts_filt = popClustering_filt_regions_afterHomologous_chr11_tarCounts %>%
filter(tarCounts >= 0.80 * max(tarCounts) |
s_Sample %in% previousDeletionCalls$BiologicalSample)
popClustering_filt_regions_afterHomologous_chr11_sampCounts = popClustering_filt_regions_afterHomologous_chr11 %>%
group_by(p_name) %>%
summarise(sampCounts = length(unique(s_Sample)))
metaByBioSample_out = metaByBioSample %>%
left_join(allDeletionTypeMeta %>%
select(-sample, -ExperimentSample) %>%
rename(sample = BiologicalSample))
write_tsv(metaByBioSample_out, "metaByBioSample_outwithHrpCalls.tab.txt")
write_tsv(popClustering_filt_regions_afterHomologous_chr11 %>%
filter(s_Sample %in% popClustering_filt_regions_afterHomologous_chr11_tarCounts_filt$s_Sample) %>%
group_by() %>%
select(s_Sample, p_name, h_popUID, c_AveragedFrac),
"popClustering_filt_regions_afterHomologous_chr11.tab.txt.gz")elucidator doPairwiseComparisonOnHapsSharingDev --tableFnp popClustering_filt_regions_afterHomologous_chr11.tab.txt.gz --sampleCol s_Sample --targetNameCol p_name --popIDCol h_popUID --relAbundCol c_AveragedFrac --numThreads 14 --dout pairwiseComps_regions_afterHomologous_chr11 --verbose --overWriteDir --metaFnp metaByBioSample_outwithHrpCalls.tab.txt --metaFieldsToCalcPopDiffs country,region,secondaryRegion,HRP3_deletionPattern --writeOutDistMatrices#jacardDist = readr::read_tsv("pairwiseComps_regions_afterHomologous_chr11/percOfTarSharingAtLeastOneHap.tab.txt.gz", col_names = F)
jacardDist = readr::read_tsv("pairwiseComps_regions_afterHomologous_chr11/jacardByHapsTarShared.tab.txt.gz", col_names = F)
jacardDistSamps = readr::read_tsv("pairwiseComps_regions_afterHomologous_chr11/sampleNames.tab.txt", col_names = "samples")
colnames(jacardDist) = jacardDistSamps$samples
jacardDist$sample = jacardDistSamps$samples
# jacardDist_filt = jacardDist[jacardDist$sample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample,c(allDeletionTypeMeta_hrp3_pat1$BiologicalSample, "sample")]
# jacardDist_gat = jacardDist_filt %>%
# gather(otherSample, index,1:(ncol(.) - 1))
jacardDist_gat = jacardDist %>%
gather(otherSample, index,1:(ncol(.) - 1))
jacardDist_gat_filt = jacardDist_gat %>%
filter(sample %fin% allDeletionTypeMeta_hrp3_pat1$BiologicalSample,
otherSample %fin% allDeletionTypeMeta_hrp3_pat1$BiologicalSample)
jacardDist_gat_filt_sp = jacardDist_gat_filt %>%
spread(otherSample, index)
jacardDist_gat_filt_sp_mat = as.matrix(jacardDist_gat_filt_sp[,2:ncol(jacardDist_gat_filt_sp)])
rownames(jacardDist_gat_filt_sp_mat) = jacardDist_gat_filt_sp$samplejacardDist_gat_filt_sp_mat_pat1 = jacardDist_gat_filt_sp_mat
# get data just for variable regions (e.g., minPopSize = 2)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11, minPopSize = 2)
# cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample))) %>%
group_by() %>%
filter(sampleCount >= 0.99*max(sampleCount)) %>% # get just targets with high sample coverage otherwise clustering will be by missingness
group_by(s_Sample, p_name) %>%
#filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist)
# get clustering based on the jacard distance too for reference
jacardDist_gat_filt_sp_mat_pat1_hc = hclust(dist(jacardDist_gat_filt_sp_mat_pat1))
k_groups = 42;
h_groups = 1.1;
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups = cutree(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust, k = k_groups)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend <- as.dendrogram(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend <- color_labels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend, k = k_groups)
plot(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups = cutree(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust, h = h_groups)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend <- as.dendrogram(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend <- color_labels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend, h = h_groups)
plot(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend)
quartz_off_screen
2 jacardDist_gat_filt_sp_mat_pat1_hc_groups = cutree(jacardDist_gat_filt_sp_mat_pat1_hc, k = k_groups)
jacardDist_gat_filt_sp_mat_pat1_hc_dend <- as.dendrogram(jacardDist_gat_filt_sp_mat_pat1_hc)
jacardDist_gat_filt_sp_mat_pat1_hc_dend <- color_labels(jacardDist_gat_filt_sp_mat_pat1_hc_dend, k = k_groups)
plot(jacardDist_gat_filt_sp_mat_pat1_hc_dend)
quartz_off_screen
2 popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df = tibble(
BiologicalSample = names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups),
hcclust = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups
) %>%
# mutate(BiologicalSample =factor(BiologicalSample, levels = levels(meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering$BiologicalSample))) %>%
group_by(hcclust) %>%
mutate(hcclustSize = n())
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_biggerGroups = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>% filter(hcclustSize != 1) %>% select(hcclust) %>% unique()
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_singletonGroups = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>% filter(hcclustSize == 1) %>% select(hcclust) %>% unique()
newscheme <- iwanthue(seed = 626, force_init = TRUE); newscheme$hex(8)[1] "#ba9d50" "#7947b8" "#8fcf52" "#c25191" "#78b795" "#c35540" "#979bc2" "#4c3c3d"# nonSingletonGroupsColors = createColorListFromDf(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_biggerGroups)$hcclust
nonSingletonGroupsColors = newscheme$hex(nrow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_biggerGroups))
names(nonSingletonGroupsColors) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_biggerGroups$hcclust
nonSingletonGroupsColors_singleton = rep("grey71", nrow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_singletonGroups))
names(nonSingletonGroupsColors_singleton) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_singletonGroups$hcclust
haploGroupColors = c(nonSingletonGroupsColors, nonSingletonGroupsColors_singleton)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
select(hcclust, hcclustSize) %>%
ungroup() %>%
unique() %>%
arrange(desc(hcclustSize)) %>%
mutate(hcclust = as.character(hcclust),
newClusterName = row_number()) %>%
left_join(tibble(
hcclust = names(haploGroupColors),
colors = unname(haploGroupColors)
)) %>%
mutate(Chr11DupHapCluster = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, pad = "0", width = 2)))
newHaploGroupColors = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts$colors
names(newHaploGroupColors)= popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts$newClusterName
newHaploGroupWithSingletColors = c(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts$colors[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts$hcclustSize > 1],
"grey77")
names(newHaploGroupWithSingletColors)= c(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts$Chr11DupHapCluster[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts$hcclustSize > 1],
"singlet")
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts %>%
mutate(hcclust = as.integer(hcclust)))
write_tsv(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df, "popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df.tsv")library(circlize)
#col_fun = colorRamp2(c(0, 0.5, 1), c(heat.colors(3)))
col_fun = colorRamp2(c(min(jacardDist_gat_filt_sp_mat), min(jacardDist_gat_filt_sp_mat) + (1-min(jacardDist_gat_filt_sp_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
jacardDist_gat_filt_sp_mat_noLabs = jacardDist_gat_filt_sp_mat
jacardDist_gat_filt_sp_mat_pat1 = jacardDist_gat_filt_sp_mat
meta_preferredSample = metaSelected %>%
filter(PreferredSample)
metaSelected_hrp3_pat1 = meta_preferredSample[match(rownames(jacardDist_gat_filt_sp_mat), meta_preferredSample$BiologicalSample), ]%>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
select(BiologicalSample, Chr11DupHapCluster))
metaSelected_hrp3_pat1 = metaSelected_hrp3_pat1 %>%
mutate(PerfectChr11Copy = BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies$s_Sample)
rownames(jacardDist_gat_filt_sp_mat_noLabs) = NULL
colnames(jacardDist_gat_filt_sp_mat_noLabs) = NULL
RowLabs = metaSelected_hrp3_pat1$BiologicalSample
RowLabs[metaSelected_hrp3_pat1$site != "LabIsolate" | is.na(metaSelected_hrp3_pat1$site)] = ""
ColLabs = metaSelected_hrp3_pat1$BiologicalSample
ColLabs[metaSelected_hrp3_pat1$site != "LabIsolate" | is.na(metaSelected_hrp3_pat1$site)] = ""
#RowLabs[metaSelected$country != "Ethiopia"] = ""
rownames(jacardDist_gat_filt_sp_mat_noLabs) = RowLabs
colnames(jacardDist_gat_filt_sp_mat_noLabs) = ColLabs
rowAnnoDf = metaSelected_hrp3_pat1[,c("hrpCall", "PerfectChr11Copy", "country", "region", "secondaryRegion", "Chr11DupHapCluster")] %>% rename(continent = secondaryRegion) %>% as.data.frame()
annotationTextSize = 25 ;annotationTitleTextSize = 20;
rowAnnoColors[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors,
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
show_legend = F,
gp = gpar(col = "grey10")
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
col = rowAnnoColors,
gp = gpar(col = "grey10")
)
haptype_hrp3_pat1HeatMap = Heatmap(
jacardDist_gat_filt_sp_mat_noLabs,
cluster_columns = T,
col = col_fun,
name = "JacardIndex",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "JacardIndex"
)
)
)Jacard index of the duplicated region on chromosome 11, jacard of 1 means complete agreement between samples on this region which 0 would be no haplotypes shared in this region. Additional meta data of the samples is shown on top and to the right including country/region, and the hrp2/3 calls, whether the the Chr11 that has been duplicated is a perfect copy or not.
It appears the African samples and South American samples, while related within continent, are not very closely related to each other.
draw(haptype_hrp3_pat1HeatMap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")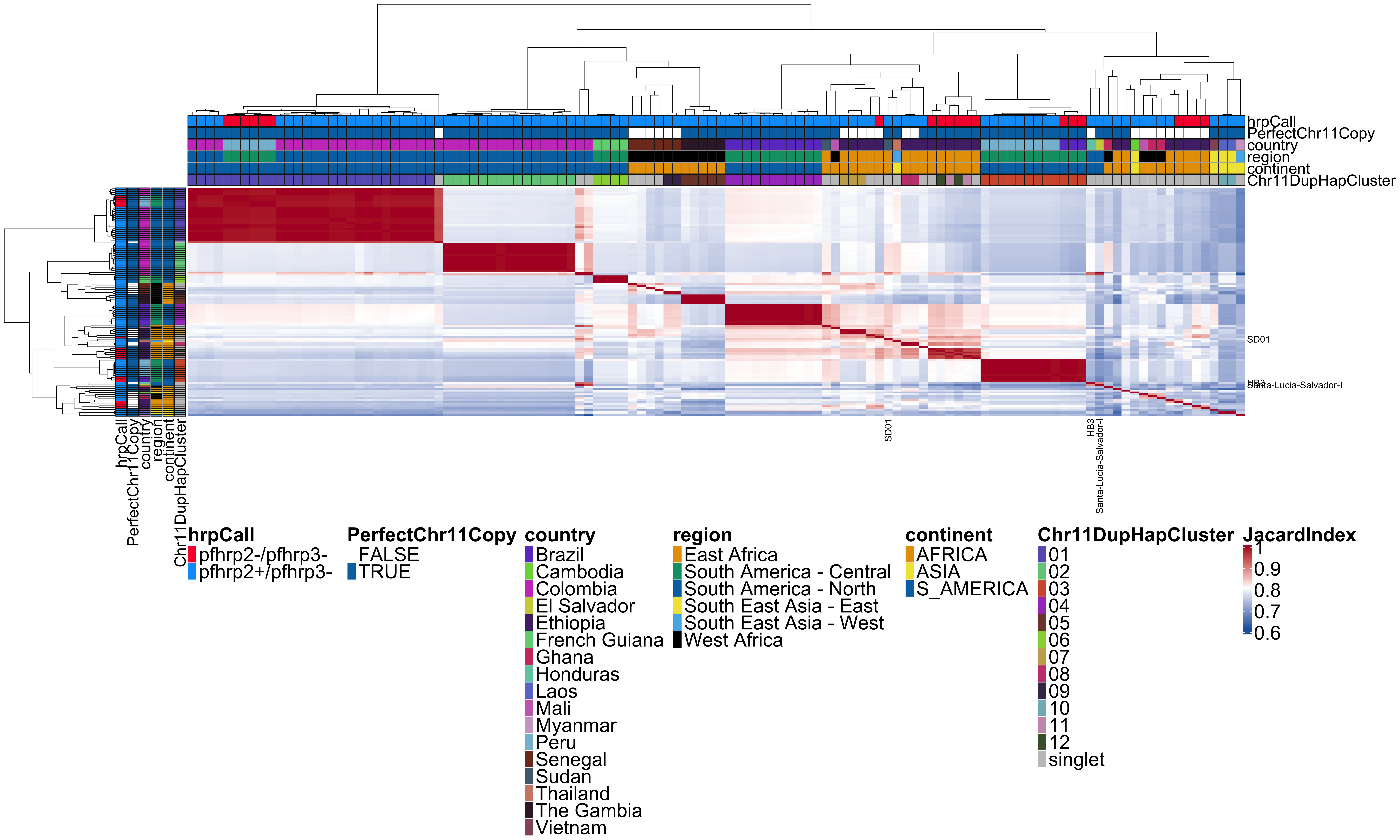
quartz_off_screen
2 Below will get the samples that have a chromosome 11 that is similar to the 13-11++ samples (Deleted hrp3, duplicated sub-telomeric chr11 segment). These new similar samples will be regardless of hrp2/3 deletion status. This will show if the chr11 that has been duplicated is circulating in the general population or is only associated with the samples with HRP3 deletion and chr11 duplication.
metaByBioSample_fieldOrIsolate = metaByBioSample %>%
filter(IsFieldSample | "LabIsolate" == site)
jacardDist_gat_filt_forOtherSimilarToPat1 = jacardDist_gat %>%
filter(sample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample |
otherSample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample ) %>%
filter(sample %in% metaByBioSample_fieldOrIsolate$sample,
otherSample %in% metaByBioSample_fieldOrIsolate$sample) %>%
filter(sample %in% samplesCovered$BiologicalSample,
otherSample %in% samplesCovered$BiologicalSample) %>%
# filter(index > 0.99)
filter(index > 0.98)
simSamples = c(unique(c(jacardDist_gat_filt_forOtherSimilarToPat1$sample, jacardDist_gat_filt_forOtherSimilarToPat1$otherSample, allDeletionTypeMeta_hrp3_pat1$BiologicalSample)))
simSamples = simSamples[simSamples != "FCR3"]
jacardDist_gat_filt_simToPat1 = jacardDist_gat %>%
filter(sample %in% simSamples,
otherSample %in% simSamples) %>%
mutate(index = ifelse(is.nan(index), 0, index))
jacardDist_gat_filt_simToPat1_sp = jacardDist_gat_filt_simToPat1 %>%
spread(otherSample, index)
jacardDist_gat_filt_simToPat1_sp_mat = as.matrix(jacardDist_gat_filt_simToPat1_sp[,2:ncol(jacardDist_gat_filt_simToPat1_sp)])
rownames(jacardDist_gat_filt_simToPat1_sp_mat) = jacardDist_gat_filt_simToPat1_sp$samplelibrary(circlize)
#['#b2182b','#d6604d','#f4a582','#fddbc7','#f7f7f7','#d1e5f0','#92c5de','#4393c3','#2166ac']
col_fun = colorRamp2(c(min(jacardDist_gat_filt_simToPat1_sp_mat), min(jacardDist_gat_filt_simToPat1_sp_mat) + (1-min(jacardDist_gat_filt_simToPat1_sp_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
jacardDist_gat_filt_simToPat1_sp_mat_noLabs = jacardDist_gat_filt_simToPat1_sp_mat
meta_preferredSample = meta %>%
filter(PreferredSample)
metaSelected_hrp3_pat1 = meta_preferredSample[match(rownames(jacardDist_gat_filt_simToPat1_sp_mat), meta_preferredSample$BiologicalSample), ] %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
select(BiologicalSample, Chr11DupHapCluster))
sample_metadata_withAllDeletionCalls = readr::read_tsv("sample_metadata_withAllDeletionCalls.tsv")
metaSelected_hrp3_pat1 = metaSelected_hrp3_pat1 %>%
left_join(sample_metadata_withAllDeletionCalls %>%
rename(BiologicalSample = sample) %>%
select(BiologicalSample, Pattern)) %>%
mutate(PerfectChr11Copy = case_when(
BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies$s_Sample ~ T,
BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum$s_Sample ~ F,
T ~ NA
))
# %>%
# mutate(hrpCall = ifelse(BiologicalSample %in% previousDeletionCalls$BiologicalSample, hrpCall, "unknown"))
rownames(jacardDist_gat_filt_simToPat1_sp_mat_noLabs) = NULL
colnames(jacardDist_gat_filt_simToPat1_sp_mat_noLabs) = NULL
RowLabs = metaSelected_hrp3_pat1$BiologicalSample
RowLabs[metaSelected_hrp3_pat1$site != "LabIsolate" | is.na(metaSelected_hrp3_pat1$site)] = ""
ColLabs = metaSelected_hrp3_pat1$BiologicalSample
ColLabs[metaSelected_hrp3_pat1$site != "LabIsolate" | is.na(metaSelected_hrp3_pat1$site)] = ""
#RowLabs[metaSelected$country != "Ethiopia"] = ""
rownames(jacardDist_gat_filt_simToPat1_sp_mat_noLabs) = RowLabs
colnames(jacardDist_gat_filt_simToPat1_sp_mat_noLabs) = ColLabs
rowAnnoDf = metaSelected_hrp3_pat1[,c("Pattern", "hrpCall", "PerfectChr11Copy", "country", "region", "secondaryRegion", "Chr11DupHapCluster")] %>% rename(continent = secondaryRegion) %>% as.data.frame()
temp_rowAnnoColors = createColorListFromDf(rowAnnoDf)
temp_rowAnnoColors[["hrpCall"]] = pfhrpsCallColors
temp_rowAnnoColors[["continent"]] = continentColors
temp_rowAnnoColors[["region"]] = rowAnnoColors$region
temp_rowAnnoColors[["Pattern"]] = rowAnnoColorsMod_hrp3DeletionPattern
temp_rowAnnoColors[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
annotationTextSize = 25 ;annotationTitleTextSize = 20;
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = temp_rowAnnoColors,
show_legend = F,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = c("#99999900")
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = temp_rowAnnoColors,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = c("#99999900")
)
haptype_simTo_hrp3_pat1HeatMap = Heatmap(
jacardDist_gat_filt_simToPat1_sp_mat_noLabs,
cluster_columns = T,
col = col_fun,
name = "JacardIndex",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "JacardIndex"
)
)
)Below will get the samples that have a chromosome 11 that is similar to the 13-11++ samples. These new similar samples will be regardless of hrp2/3 deletion status. This will show if the chr11 that has been duplicated is circulating in the general population or is only associated with the samples with HRP3 deletion and chr11 duplication.
It appears that the duplicated chromosome 11 is circulating fairly commonly among South American samples that don’t have HRP3 deletion while there doesn’t appear to be any of the duplicated chr11 circulating in the African population (though could be a high diversity vs low diversity bias and/or sampling biases given the drastic differences in malaria dynamics in the two continents).
draw(haptype_simTo_hrp3_pat1HeatMap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
Plotting out the variation at the duplicated region, coloring haplotypes by their abundance rank, this visualization will allow interpretation of how similar these haplotypes are here and what the copy looks like within sample (e.g. perfect copy vs variation and how much variation )
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11, minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample))) %>%
group_by() %>%
filter(sampleCount >= 0.99*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
#filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_dist)
jacardDist_gat_filt_sp_mat_pat1_hc = hclust(dist(jacardDist_gat_filt_sp_mat_pat1))
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_dist_hclust$order])) %>%
# levels = rownames(jacardDist_gat_filt_sp_mat_pat1)[jacardDist_gat_filt_sp_mat_pat1_hc$order])) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name),
expand = c(0,0))+
scale_y_continuous(expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$s_Sample) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
select(BiologicalSample, Chr11DupHapCluster)) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_dist_hclust$order])) %>%
mutate(popid= factor(popid))popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1 = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
# levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry$p_name),
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry$p_name)))
),
expand = c(0,0)) +
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1 +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapCluster)), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep)+
scale_fill_manual("Chr11DupHapCluster", values = newHaploGroupWithSingletColors, labels = names(newHaploGroupWithSingletColors),
breaks = names(newHaploGroupWithSingletColors)) +
guides(fill = guide_legend(nrow = 4)) The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype. Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal (and in this instance would mean the copy on chr11 and chr13 is not a perfect copy).
regions_afterHomologous_chr11_filt = regions_afterHomologous_chr11 %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name)))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1 +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -5,
fill = description),
data = regions_afterHomologous_chr11_filt, color = "black") +
scale_fill_manual("Genes\nDescription", values = descriptionColors,
guide = guide_legend(nrow = 5)) +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1)
quartz_off_screen
2 popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3 = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11, minPopSize = 2)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3 %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample))) %>%
group_by() %>%
filter(sampleCount >= 0.99*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
#filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3 %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_dist_hclust$order])) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$p_name),
expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3 = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$s_Sample) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
select(BiologicalSample, Chr11DupHapCluster)) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3 %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_dist_hclust$order])) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(limits = c(-30, max(c(-9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$p_name))))),
breaks = c(-9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$p_name))),
labels = c("HaploGroup", "continent", levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$p_name)),
expand = c(0,0))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 4)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 4)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapCluster)), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep)+
scale_fill_manual("Chr11DupHapCluster", values = newHaploGroupWithSingletColors, labels = names(newHaploGroupWithSingletColors),
breaks = names(newHaploGroupWithSingletColors)) +
guides(fill = guide_legend(nrow = 4))
regions_afterHomologous_chr11_filt = regions_afterHomologous_chr11 %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$p_name)))
yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$s_Sample)
yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3[yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 %!in% c("HB3", "Santa-Lucia-Salvador-I", "SD01")] = ""
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 +
scale_y_continuous(labels = yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3,
breaks = 1:length(yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3),
expand = c(0,0)) +
theme(axis.text.x = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank(),
panel.border = element_blank(),
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3_priorToGeneInfo = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -7,
fill = description),
data = regions_afterHomologous_chr11_filt, color = "black") +
geom_text(
aes(y = as.numeric(BiologicalSample),
x = -10,
label = BiologicalSample),
hjust = 1,
#data = tibble(BiologicalSample = factor(c("HB3", "Santa-Lucia-Salvador-I", "SD01"), levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$s_Sample)))
data = tibble(BiologicalSample = factor(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$s_Sample, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$s_Sample)))
) +
scale_fill_manual("Genes\nDescription", values = descriptionColors,
guide = guide_legend(nrow = 4)) The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype. Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal (and in this instance would mean the copy on chr11 and chr13 is not a perfect copy).
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3)
quartz_off_screen
2 quartz_off_screen
2 popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
group_by(s_Sample) %>%
summarise(p_name_count = length(unique(p_name)),
p_name_meanCOI = mean(uniqHaps)) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
rename(s_Sample = BiologicalSample))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness %>%
#filter(s_Sample %!in% c("HB3", "QV0040-C", "IGS-CBD-010")) %>%
#filter(hcclustSize > 2, newClusterName != 9) %>%
#filter(hcclustSize > 1, newClusterName != 9) %>%
filter(hcclustSize > 1) %>%
arrange(desc(p_name_count), p_name_meanCOI) %>%
group_by(newClusterName) %>%
mutate(groupID = row_number()) %>%
filter(groupID == 1) %>%
left_join(meta_preferredSample %>%
select(BiologicalSample, secondaryRegion) %>%
rename(s_Sample = BiologicalSample))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt %>%
mutate(secondaryRegion = factor(secondaryRegion, levels = c("S_AMERICA", "AFRICA", "ASIA"))) %>%
arrange(secondaryRegion, desc(hcclustSize)) %>%
mutate(s_Sample = factor(s_Sample, levels = .$s_Sample))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4 = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
filter(s_Sample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt$s_Sample), minPopSize = 2)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4 %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample))) %>%
group_by() %>%
filter(sampleCount >= 0.99*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist)
jacardDist_gat_filt_sp_mat_pat1_hc = hclust(dist(jacardDist_gat_filt_sp_mat_pat1))
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4 %>%
mutate(s_Sample = factor(s_Sample,
levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt$s_Sample))) %>%
# levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust$order])) %>%
# levels = rownames(jacardDist_gat_filt_sp_mat_pat1)[jacardDist_gat_filt_sp_mat_pat1_hc$order])) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$p_name),
expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4 = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$s_Sample) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4 %>%
mutate(s_Sample = factor(s_Sample,
levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt$s_Sample))) %>%
# mutate(s_Sample = factor(s_Sample,
# levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust$order])) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(limits = c(-30, max(c(-9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry$p_name))))),
breaks = c(-9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry$p_name))),
labels = c("HaploGroup", "continent", levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry$p_name)),
expand = c(0,0))
k_groups = nrow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt);
h_groups = 1.1;
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups = cutree(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust, k = k_groups)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend <- as.dendrogram(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend <- color_labels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend, k = k_groups)
plot(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups = cutree(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust, h = h_groups)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend <- as.dendrogram(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend <- color_labels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend, h = h_groups)
plot(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend)
jacardDist_gat_filt_sp_mat_pat1_hc_groups = cutree(jacardDist_gat_filt_sp_mat_pat1_hc, k = k_groups)
jacardDist_gat_filt_sp_mat_pat1_hc_dend <- as.dendrogram(jacardDist_gat_filt_sp_mat_pat1_hc)
jacardDist_gat_filt_sp_mat_pat1_hc_dend <- color_labels(jacardDist_gat_filt_sp_mat_pat1_hc_dend, k = k_groups)
plot(jacardDist_gat_filt_sp_mat_pat1_hc_dend)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df = tibble(
BiologicalSample = names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups),
hcclust = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups
) %>%
mutate(BiologicalSample =factor(BiologicalSample, levels = levels(meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$BiologicalSample))) %>%
group_by(hcclust) %>%
mutate(hcclustSize = n())
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_biggerGroups = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df %>% filter(hcclustSize != 1) %>% select(hcclust) %>% unique()
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_singletonGroups = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df %>% filter(hcclustSize == 1) %>% select(hcclust) %>% unique()
nonSingletonGroupsColors = scheme$hex(nrow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df %>% select(hcclust) %>% unique()))
names(nonSingletonGroupsColors) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_biggerGroups$hcclust
nonSingletonGroupsColors_singleton = scheme$hex(nrow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df %>% select(hcclust) %>% unique()))
names(nonSingletonGroupsColors_singleton) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_singletonGroups$hcclust
haploGroupColors = c(nonSingletonGroupsColors, nonSingletonGroupsColors_singleton)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_groupCounts = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df %>%
select(hcclust, hcclustSize) %>%
ungroup() %>%
unique() %>%
arrange(desc(hcclustSize)) %>%
mutate(hcclust = as.character(hcclust),newClusterName = row_number()) %>%
left_join(tibble(
hcclust = names(haploGroupColors),
colors = unname(haploGroupColors)
))
newHaploGroupColors = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_groupCounts$colors
names(newHaploGroupColors)= popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_groupCounts$newClusterName
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df %>%
left_join(
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_groupCounts %>%
mutate(hcclust = as.integer(hcclust))
) %>%
left_join(
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>% ungroup() %>% select(BiologicalSample, hcclustSize) %>% rename(originalGroupSize = hcclustSize)
) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$BiologicalSample)))
# jacardDist_gat_filt_sp_mat_pat1_hc_groups_df = tibble(
# BiologicalSample = names(jacardDist_gat_filt_sp_mat_pat1_hc_groups),
# hcclust = jacardDist_gat_filt_sp_mat_pat1_hc_groups
# ) %>%
# mutate(BiologicalSample =factor(BiologicalSample, levels = levels(meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$BiologicalSample)))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))]) ) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.3,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 4)) +
# ggnewscale::new_scale_fill() +
# geom_rect(aes(xmin= -5, xmax = -9.5,
# ymin = as.numeric(BiologicalSample) - 0.5,
# ymax = as.numeric(BiologicalSample) + 0.3,
# fill = factor(newClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df)+
# # fill = factor(hcclust)), color = "black", data = jacardDist_gat_filt_sp_mat_pat1_hc_groups_df)+
# # scale_fill_manual("HaploGroup", values = scheme$hex(length(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups)))) +
# #scale_fill_manual("Chr11DupHapCluster", values = haploGroupColors, labels = names(haploGroupColors), breaks = names(haploGroupColors)) +
# scale_fill_manual("Chr11DupHapCluster", values = newHaploGroupColors, labels = names(newHaploGroupColors),
# breaks = names(newHaploGroupColors)) +
geom_text(aes(
x = -9.5,
y = as.numeric(BiologicalSample) - 0.5 + 0.4,
label = paste0("n=", originalGroupSize)
), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df)+
guides(fill = guide_legend(nrow = 4))
regions_afterHomologous_chr11_filt = regions_afterHomologous_chr11 %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$p_name)))
yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry$s_Sample)
# yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4[yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 %!in% c("HB3", "Santa-Lucia-Salvador-I", "SD01")] = ""
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 +
scale_y_continuous(labels = yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4,
breaks = 1:length(yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4),
expand = c(0,0)) +
theme(axis.text.x = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank(),
panel.border = element_blank(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black", linewidth = 1)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4_priorToGeneInfo = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -1,
fill = description),
data = regions_afterHomologous_chr11_filt, color = "black") +
geom_text(
aes(y = as.numeric(BiologicalSample),
x = -10,
label = BiologicalSample),
hjust = 1,
data = tibble(BiologicalSample = factor(c("HB3", "Santa-Lucia-Salvador-I", "SD01"), levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry$s_Sample)))
) +
scale_fill_manual("Genes\nDescription", values = descriptionColors,
guide = guide_legend(nrow = 4)) The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype. Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal (and in this instance would mean the copy on chr11 and chr13 is not a perfect copy).
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4)
quartz_off_screen
2 quartz_off_screen
2 popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
filter(s_Sample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies$s_Sample) , minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample))) %>%
group_by() %>%
filter(sampleCount > 0.9*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_dist_hclust$order]))%>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$p_name),
expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$s_Sample) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_dist_hclust$order])) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry$p_name)))
# levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry$p_name)
),
expand = c(0,0)) +
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies =
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$s_Sample) %>%
mutate(BiologicalSample = factor(as.character(BiologicalSample), levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$s_Sample))) %>%
mutate(Chr11DupHapClusterName = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, width = 2, pad = "0")))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColorsDf = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies %>%
select(Chr11DupHapClusterName, colors) %>%
unique() %>%
arrange(Chr11DupHapClusterName)
#
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColors = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColorsDf$colors
names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColors) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColorsDf$Chr11DupHapClusterName
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies)+
# fill = factor(hcclust)), color = "black", data = jacardDist_gat_filt_sp_mat_pat1_hc_groups_df)+
# scale_fill_manual("HaploGroup", values = scheme$hex(length(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups)))) +
#scale_fill_manual("Chr11DupHapCluster", values = haploGroupColors, labels = names(haploGroupColors), breaks = names(haploGroupColors)) +
scale_fill_manual("Chr11DupHapCluster", values = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColors, labels = names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColors),
breaks = names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColors)) +
guides(fill = guide_legend(nrow = 4)) The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal (and in this instance would mean the copy on chr11 and chr13 is not a perfect copy)
regions_afterHomologous_chr11_filt = regions_afterHomologous_chr11 %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name)))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -5,
fill = description),
data = regions_afterHomologous_chr11_filt, color = "black") +
scale_fill_manual("Genes\nDescription", values = descriptionColors,
guide = guide_legend(nrow = 5)) +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot)
quartz_off_screen
2 popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
filter(s_Sample %!in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies$s_Sample) , minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies %>%
group_by() %>%
filter(samp_n > 0.9*max(samp_n)) %>%
group_by(s_Sample, p_name) %>%
#filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_dist)
nameOrderFromforClustering = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust$order]
orderForDivergentCopy = nameOrderFromforClustering[nameOrderFromforClustering %in% rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_mat)]
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies %>%
mutate(s_Sample = factor(s_Sample,
# levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_dist_hclust$order]))%>%
levels = orderForDivergentCopy)) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$p_name),
expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$s_Sample) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies %>%
mutate(s_Sample = factor(s_Sample,
# levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_dist_hclust$order])) %>%
levels = orderForDivergentCopy)) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies =
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$s_Sample) %>%
mutate(BiologicalSample = factor(as.character(BiologicalSample), levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$s_Sample))) %>%
mutate(Chr11DupHapClusterName = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, width = 2, pad = "0"))) %>%
arrange(BiologicalSample)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColorsDf = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies %>%
select(Chr11DupHapClusterName, colors) %>%
unique() %>%
arrange(Chr11DupHapClusterName)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColors = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColorsDf$colors
names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColors) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColorsDf$Chr11DupHapClusterName
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry$p_name)))
# levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry$p_name)
),
expand = c(0,0))+
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot+
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies)+
# fill = factor(hcclust)), color = "black", data = jacardDist_gat_filt_sp_mat_pat1_hc_groups_df)+
# scale_fill_manual("HaploGroup", values = scheme$hex(length(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups)))) +
#scale_fill_manual("Chr11DupHapCluster", values = haploGroupColors, labels = names(haploGroupColors), breaks = names(haploGroupColors)) +
scale_fill_manual("Chr11DupHapCluster", values = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColors, labels = names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColors),
breaks = names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColors)) +
guides(fill = guide_legend(nrow = 4)) The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal (and in this instance would mean the copy on chr11 and chr13 is not a perfect copy)
regions_afterHomologous_chr11_filt = regions_afterHomologous_chr11 %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name)))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -1,
fill = description),
data = regions_afterHomologous_chr11_filt, color = "black") +
scale_fill_manual("Genes\nDescription", values = descriptionColors,
guide = guide_legend(nrow = 5)) +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot)
quartz_off_screen
2 popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
filter(s_Sample %in% c("HB3", "SD01", "Santa-Lucia-Salvador-I")) , minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample)))%>%
group_by() %>%
filter(sampleCount > 0.9*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_dist_hclust$order]))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$p_name)),
# labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$p_name),
labels = rep("", length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$p_name))),
expand = c(0,0))
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$popid))
previousColors["-1"] = "grey0";
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates%>%
mutate(popid= factor(popid)), colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$p_name)),
# labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$p_name),
labels = rep("", length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$p_name))),
expand = c(0,0))The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal (and in this instance would mean the copy on chr11 and chr13 is not a perfect copy)
It appears that SD01 and Santa-Lucia-Salvador-I have perfect copies of chr11 on chr11 and chr13 while HB3 has a divergent copy (which is confirmed with the nanopore assembly)
Interestingly enough, the Santa-Lucia-Salvador-I chr11 duplicated region appears to be one of the chr11 in HB3.
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 1)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -1,
fill = description),
data = regions_afterHomologous_chr11_filt, color = "black") +
scale_fill_manual("Genes\nDescription", values = descriptionColors,
guide = guide_legend(nrow = 5)) +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30),
axis.text.y = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot)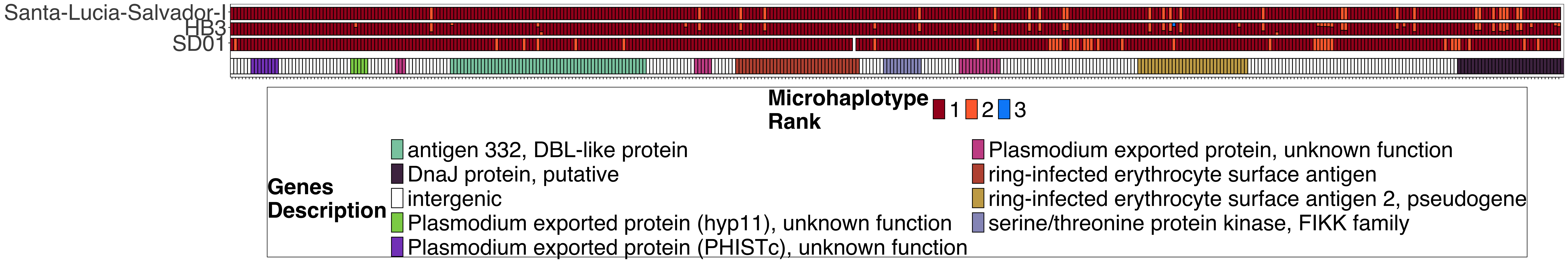
The data on the 15.2kb duplicated region between chromosome 11 and 13.
excludeRegions = c("Pf3D7_11_v3-1919633-1920323-for__var-6",
"Pf3D7_11_v3-1920483-1921173-for__var-3",
"Pf3D7_11_v3-1920483-1921173-for__var-4",
"Pf3D7_11_v3-1920483-1921173-for__var-5",
"Pf3D7_11_v3-1920483-1921173-for__var-6",
"Pf3D7_11_v3-1920483-1921173-for__var-7",
"Pf3D7_11_v3-1928369-1928869-for__var-3",
"Pf3D7_11_v3-1928619-1929119-for__var-3")
regions_homologousRegion = regions %>%
filter("shared" == homologousRegion) %>%
filter(`#chrom` == "Pf3D7_11_v3") %>%
filter(name %!in% excludeRegions)
popClustering_filt_hrp3_pat1_regions_homologousRegion = popClustering_filt_hrp3_pat1 %>%
filter(p_name %in% regions_homologousRegion$genomicID)
popClustering_filt_hrp3_pat1_regions_homologousRegion = popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
group_by(s_Sample, p_name) %>%
mutate(uniqHaps= n())
popClustering_filt_hrp3_pat1_regions_homologousRegion_uniqSum = popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
group_by(s_Sample) %>%
mutate(targets = length(unique(genomicID))) %>%
group_by(s_Sample, targets, uniqHaps) %>%
count() %>%
mutate(freq = n/targets)
popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum = popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
mutate(marker = uniqHaps == 1) %>%
group_by(s_Sample) %>%
summarise(conserved = sum(marker),
targets = length(unique(genomicID))) %>%
mutate(conservedID = conserved/targets)
conservedCutOff = 0.99
popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_closeToPerfectCopies = popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum %>%
filter(conservedID > conservedCutOff)
popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_cutOff = popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
group_by() %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)
popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_cutOffByRegion = popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
left_join(metaByBioSample %>%
rename(s_Sample = sample)) %>%
group_by(region) %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)
popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_cutOffByContinent = popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
left_join(metaByBioSample %>%
rename(s_Sample = sample)) %>%
group_by(secondaryRegion) %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)The number of samples with perfect copies
create_dt(popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_cutOff)The number of samples with perfect copies broken down by regions
create_dt(popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_cutOffByRegion)The number of samples with perfect copies broken down by continent.
create_dt(popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_cutOffByContinent) The breakdown of level of divergence in the samples with divergent samples.
create_dt(popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_meanId)Calculating the population of the haplotypes of the shared region on chr 11/chr13
popClustering_filt_regions_homologousRegion = popClustering %>%
filter(genomicID %!in% erroneousRegions) %>%
filter(p_name %in% regions_homologousRegion$genomicID)
popClustering_filt_regions_homologousRegion_tarCounts = popClustering_filt_regions_homologousRegion %>%
group_by(s_Sample) %>%
summarise(tarCounts = length(unique(p_name)))
popClustering_filt_regions_homologousRegion_tarCounts_filt = popClustering_filt_regions_homologousRegion_tarCounts %>%
filter(tarCounts >= 0.80 * max(tarCounts) |
s_Sample %in% previousDeletionCalls$BiologicalSample)
popClustering_filt_regions_homologousRegion_sampCounts = popClustering_filt_regions_homologousRegion %>%
group_by(p_name) %>%
summarise(sampCounts = length(unique(s_Sample)))
write_tsv(popClustering_filt_regions_homologousRegion %>%
filter(s_Sample %in% popClustering_filt_regions_homologousRegion_tarCounts_filt$s_Sample) %>%
group_by() %>%
select(s_Sample, p_name, h_popUID, c_AveragedFrac),
"popClustering_filt_regions_homologousRegion.tab.txt.gz")elucidator doPairwiseComparisonOnHapsSharingDev --tableFnp popClustering_filt_regions_homologousRegion.tab.txt.gz --sampleCol s_Sample --targetNameCol p_name --popIDCol h_popUID --relAbundCol c_AveragedFrac --numThreads 14 --dout pairwiseComps_regions_homologousRegion --verbose --overWriteDir --metaFnp metaByBioSample_outwithHrpCalls.tab.txt --metaFieldsToCalcPopDiffs country,region,secondaryRegion,HRP3_deletionPattern --writeOutDistMatrices#jacardDist = readr::read_tsv("pairwiseComps_regions_homologousRegion/percOfTarSharingAtLeastOneHap.tab.txt.gz", col_names = F)
jacardDist_homologousRegion = readr::read_tsv("pairwiseComps_regions_homologousRegion/jacardByHapsTarShared.tab.txt.gz", col_names = F)
jacardDist_homologousRegionSamps = readr::read_tsv("pairwiseComps_regions_homologousRegion/sampleNames.tab.txt", col_names = "samples")
colnames(jacardDist_homologousRegion) = jacardDist_homologousRegionSamps$samples
jacardDist_homologousRegion$sample = jacardDist_homologousRegionSamps$samples
# jacardDist_homologousRegion_filt = jacardDist_homologousRegion %>%
# filter(sample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample)
jacardDist_homologousRegion_filt = jacardDist_homologousRegion[jacardDist_homologousRegion$sample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample,c(allDeletionTypeMeta_hrp3_pat1$BiologicalSample, "sample")]
jacardDist_homologousRegion_gat = jacardDist_homologousRegion_filt %>%
gather(otherSample, index,1:(ncol(.) - 1))
jacardDist_homologousRegion_gat_filt = jacardDist_homologousRegion_gat %>%
filter(sample %fin% allDeletionTypeMeta_hrp3_pat1$BiologicalSample,
otherSample %fin% allDeletionTypeMeta_hrp3_pat1$BiologicalSample)
jacardDist_homologousRegion_gat_filt_sp = jacardDist_homologousRegion_gat_filt %>%
spread(otherSample, index)
jacardDist_homologousRegion_gat_filt_sp_mat = as.matrix(jacardDist_homologousRegion_gat_filt_sp[,2:ncol(jacardDist_homologousRegion_gat_filt_sp)])
rownames(jacardDist_homologousRegion_gat_filt_sp_mat) = jacardDist_homologousRegion_gat_filt_sp$samplelibrary(circlize)
#col_fun = colorRamp2(c(0, 0.5, 1), c(heat.colors(3)))
col_fun = colorRamp2(c(min(jacardDist_homologousRegion_gat_filt_sp_mat), min(jacardDist_homologousRegion_gat_filt_sp_mat) + (1-min(jacardDist_homologousRegion_gat_filt_sp_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
jacardDist_homologousRegion_gat_filt_sp_mat_noLabs = jacardDist_homologousRegion_gat_filt_sp_mat
meta_preferredSample = meta %>%
filter(PreferredSample)
metaSelected_hrp3_pat1 = meta_preferredSample[match(rownames(jacardDist_homologousRegion_gat_filt_sp_mat), meta_preferredSample$BiologicalSample), ]
metaSelected_hrp3_pat1 = metaSelected_hrp3_pat1 %>%
mutate(PerfectChr11Copy = BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies$s_Sample) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
mutate(newClusterName = ifelse(hcclustSize == 1, "singlet", as.character(stringr::str_pad(newClusterName, width = 2, pad = "0")))) %>%
mutate(BiologicalSample = as.character(BiologicalSample)) %>%
select(BiologicalSample, newClusterName))
rownames(jacardDist_homologousRegion_gat_filt_sp_mat_noLabs) = NULL
colnames(jacardDist_homologousRegion_gat_filt_sp_mat_noLabs) = NULL
RowLabs = metaSelected_hrp3_pat1$BiologicalSample
RowLabs[metaSelected_hrp3_pat1$site != "LabIsolate" | is.na(metaSelected_hrp3_pat1$site)] = ""
ColLabs = metaSelected_hrp3_pat1$BiologicalSample
ColLabs[metaSelected_hrp3_pat1$site != "LabIsolate" | is.na(metaSelected_hrp3_pat1$site)] = ""
#RowLabs[metaSelected$country != "Ethiopia"] = ""
rownames(jacardDist_homologousRegion_gat_filt_sp_mat_noLabs) = RowLabs
colnames(jacardDist_homologousRegion_gat_filt_sp_mat_noLabs) = ColLabs
rowAnnoDf = metaSelected_hrp3_pat1[,c("hrpCall", "PerfectChr11Copy", "country", "region", "secondaryRegion", "newClusterName")] %>% rename(continent = secondaryRegion,
Chr11DupHapCluster = newClusterName) %>% as.data.frame()
annotationTextSize = 25 ;annotationTitleTextSize = 20;
rowAnnoColors_mod = rowAnnoColors
rowAnnoColors_mod[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
show_legend = F,
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
gp = gpar(col = "grey10")
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
gp = gpar(col = "grey10")
)
haptype_hrp3_regions_homologousRegion_pat1HeatMap = Heatmap(
jacardDist_homologousRegion_gat_filt_sp_mat_noLabs,
cluster_columns = T,
col = col_fun,
name = "JacardIndex",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "JacardIndex"
)
)
)draw(haptype_hrp3_regions_homologousRegion_pat1HeatMap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")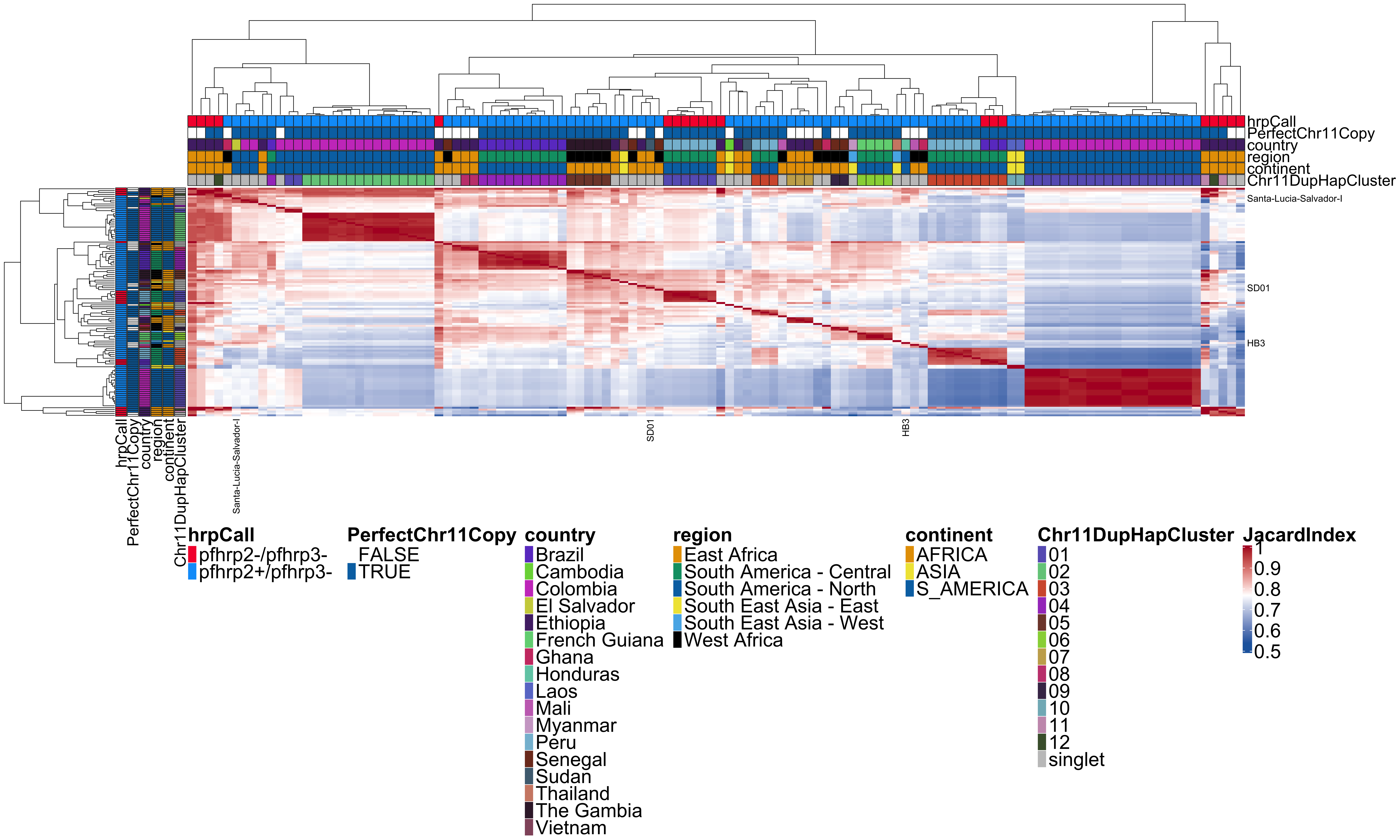
quartz_off_screen
2 Plotting out the variation at the duplicated region, coloring haplotypes by their abundance rank, this visualization will allow interpretation of how similar these haplotypes are here and what the copy looks like within sample (e.g. perfect copy vs variation and how much variation )
regions_homologousRegion = regions_homologousRegion %>%
mutate(description = case_when(
grepl("extraField0=NA", extraField0) ~ "intergenic",
T ~ gsub("\\]", "", gsub(".*description=", "", extraField0))
) )
descriptionColors_homologousRegion = scheme$hex(length(regions_homologousRegion$description %>% unique()))
names(descriptionColors_homologousRegion) = regions_homologousRegion$description %>% unique()
descriptionColors_homologousRegion["intergenic"] = c("#FF000000")
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_homologousRegion, minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample)))%>%
group_by() %>%
filter(sampleCount > 0.9*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_mat) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_mat)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_dist_hclust$order])) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$p_name),
expand = c(0,0)) +
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$s_Sample)
)
meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$s_Sample) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_dist_hclust$order])) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$p_name)))
# levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$p_name)
),
expand = c(0,0))+
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry =
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$s_Sample) %>%
mutate(BiologicalSample = factor(as.character(BiologicalSample), levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$s_Sample))) %>%
mutate(Chr11DupHapClusterName = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, width = 2, pad = "0"))) %>%
arrange(BiologicalSample)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColorsDf = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry %>%
select(Chr11DupHapClusterName, colors) %>%
unique() %>%
arrange(Chr11DupHapClusterName)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColors = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColorsDf$colors
names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColors) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColorsDf$Chr11DupHapClusterName
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry)+
# fill = factor(hcclust)), color = "black", data = jacardDist_gat_filt_sp_mat_pat1_hc_groups_df)+
# scale_fill_manual("HaploGroup", values = scheme$hex(length(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups)))) +
#scale_fill_manual("Chr11DupHapCluster", values = haploGroupColors, labels = names(haploGroupColors), breaks = names(haploGroupColors)) +
scale_fill_manual("Chr11DupHapCluster", values = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColors, labels = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColors),
breaks = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColors)) +
guides(fill = guide_legend(nrow = 4)) The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal
regions_homologousRegion_filt = regions_homologousRegion %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$p_name)))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot_final = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -10,
fill = description),
data = regions_homologousRegion_filt, color = "black") +
scale_fill_manual("Genes\nDescription", values = descriptionColors_homologousRegion,
guide = guide_legend(nrow = 2) ) +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot_final )
quartz_off_screen
2 popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
filter(s_Sample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_closeToPerfectCopies$s_Sample) , minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample)))%>%
group_by() %>%
filter(sampleCount > 0.9*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_mat) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_mat)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_dist_hclust$order]))%>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies$p_name),
expand = c(0,0))+
scale_y_continuous(expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies$s_Sample) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_dist_hclust$order])) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$p_name)))
# levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$p_name)
),
expand = c(0,0))+
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry =
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$s_Sample) %>%
mutate(BiologicalSample = factor(as.character(BiologicalSample), levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$s_Sample))) %>%
mutate(Chr11DupHapClusterName = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, width = 2, pad = "0"))) %>%
arrange(BiologicalSample)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColorsDf = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry %>%
select(Chr11DupHapClusterName, colors) %>%
unique() %>%
arrange(Chr11DupHapClusterName)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColors = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColorsDf$colors
names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColors) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColorsDf$Chr11DupHapClusterName
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot+
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry)+
# fill = factor(hcclust)), color = "black", data = jacardDist_gat_filt_sp_mat_pat1_hc_groups_df)+
# scale_fill_manual("HaploGroup", values = scheme$hex(length(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups)))) +
#scale_fill_manual("Chr11DupHapCluster", values = haploGroupColors, labels = names(haploGroupColors), breaks = names(haploGroupColors)) +
scale_fill_manual("Chr11DupHapCluster", values = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColors, labels = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColors),
breaks = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColors)) +
guides(fill = guide_legend(nrow = 4)) The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal
regions_homologousRegion_filt = regions_homologousRegion %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$p_name)))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot_final = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -1,
fill = description),
data = regions_homologousRegion_filt, color = "black") +
scale_fill_manual("Genes\nDescription", values = descriptionColors_homologousRegion,
guide = guide_legend(nrow = 2))+
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot_final)
Divergent copies of the shared region
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
filter(s_Sample %!in% popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_closeToPerfectCopies$s_Sample) , minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies %>%
group_by() %>%
filter(samp_n > 0.9*max(samp_n)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_mat) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_mat)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_dist)
nameOrderFromAll = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_dist_hclust$order]
orderForDivergentCopy = nameOrderFromAll[nameOrderFromAll %in% rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_mat)]
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies %>%
mutate(s_Sample = factor(s_Sample,
# levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_dist_hclust$order]))%>%
levels = orderForDivergentCopy)) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies$p_name),
expand = c(0,0))+
scale_y_continuous(expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies$s_Sample) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies %>%
mutate(s_Sample = factor(s_Sample,
# levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_dist_hclust$order])) %>%
levels = orderForDivergentCopy)) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$p_name)))
# levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$p_name)
),
expand = c(0,0))+
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry =
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$s_Sample) %>%
mutate(BiologicalSample = factor(as.character(BiologicalSample), levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$s_Sample))) %>%
mutate(Chr11DupHapClusterName = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, width = 2, pad = "0"))) %>%
arrange(BiologicalSample)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColorsDf = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry %>%
select(Chr11DupHapClusterName, colors) %>%
unique() %>%
arrange(Chr11DupHapClusterName)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColors = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColorsDf$colors
names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColors) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColorsDf$Chr11DupHapClusterName
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry)+
# fill = factor(hcclust)), color = "black", data = jacardDist_gat_filt_sp_mat_pat1_hc_groups_df)+
# scale_fill_manual("HaploGroup", values = scheme$hex(length(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups)))) +
#scale_fill_manual("Chr11DupHapCluster", values = haploGroupColors, labels = names(haploGroupColors), breaks = names(haploGroupColors)) +
scale_fill_manual("Chr11DupHapCluster", values = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColors, labels = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColors),
breaks = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColors)) +
guides(fill = guide_legend(nrow = 4)) The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal
regions_homologousRegion_filt = regions_homologousRegion %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$p_name)))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot_final = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -10,
fill = description),
data = regions_homologousRegion_filt, color = "black") +
scale_fill_manual(values = descriptionColors_homologousRegion,
guide = guide_legend(nrow = 2)) +
labs(fill = "Genes\nDescription") +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot_final)
The shared region of the strains with perfect chr11 copies.
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies = HaplotypeRainbows::prepForRainbow(
popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
filter(
s_Sample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies$s_Sample
),
minPopSize = 1
)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies %>%
group_by() %>%
# filter(samp_n > 0.9*max(samp_n)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_mat) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_mat)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist_hclust_orderDf =
tibble(
BiologicalSample = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist_hclust$order]
) %>%
mutate(byGenomicRegionHclustOrder = row_number())
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
arrange(newClusterName)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_select = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies$s_Sample) %>%
left_join(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist_hclust_orderDf) %>%
left_join(meta_preferredSample %>%
select(BiologicalSample, country, subRegion, region, secondaryRegion)) %>%
arrange(Chr11DupHapCluster, subRegion, country, byGenomicRegionHclustOrder)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies %>%
mutate(s_Sample = factor(s_Sample,
#levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist_hclust$order]))%>%
levels = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_select$BiologicalSample) ) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies$p_name),
expand = c(0,0))+
scale_y_continuous(expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies$s_Sample) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_select %>%
select(BiologicalSample, newClusterName))%>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies %>%
# mutate(s_Sample = factor(s_Sample,
# levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist_hclust$order])) %>%
mutate(s_Sample = factor(s_Sample,
levels = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_select$BiologicalSample)) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$p_name)))
# levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$p_name)
),
expand = c(0,0))+
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry =
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$s_Sample) %>%
mutate(BiologicalSample = factor(as.character(BiologicalSample), levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$s_Sample))) %>%
mutate(Chr11DupHapClusterName = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, width = 2, pad = "0"))) %>%
arrange(BiologicalSample)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColorsDf = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry %>%
select(Chr11DupHapClusterName, colors) %>%
unique() %>%
arrange(Chr11DupHapClusterName)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColors = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColorsDf$colors
names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColors) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColorsDf$Chr11DupHapClusterName
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry)+
scale_fill_manual("Chr11DupHapCluster", values = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColors, labels = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColors),
breaks = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColors)) +
guides(fill = guide_legend(nrow = 4)) The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal
regions_homologousRegion_filt = regions_homologousRegion %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$p_name)))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot_final = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -10,
fill = description),
data = regions_homologousRegion_filt, color = "black") +
scale_fill_manual(values = descriptionColors_homologousRegion,
guide = guide_legend(nrow = 2)) +
labs(fill = "Genes\nDescription") +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot_final)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
filter(s_Sample %in% c("HB3", "SD01", "Santa-Lucia-Salvador-I")) , minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample)))%>%
group_by()%>%
filter(sampleCount > 0.9*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_mat) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_mat)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_dist_hclust$order]))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name)),
#labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name),
labels = rep("", length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name))),
expand = c(0,0))+
scale_y_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$s_Sample),
expand = c(0,0))
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$popid))
previousColors["-1"] = "grey0";
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates %>%
mutate(popid= factor(popid)), colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name)),
#labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name),
labels = rep("", length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name))),
expand = c(0,0))+
scale_y_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$s_Sample),
expand = c(0,0))The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal
regions_homologousRegion_filt = regions_homologousRegion %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name)))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot_final = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 1)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -1,
fill = description),
data = regions_homologousRegion_filt, color = "black") +
scale_fill_manual(values = descriptionColors_homologousRegion,
guide = guide_legend(nrow = 2)) +
labs(fill = "Genes\nDescription") +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30),
axis.text.y = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot_final)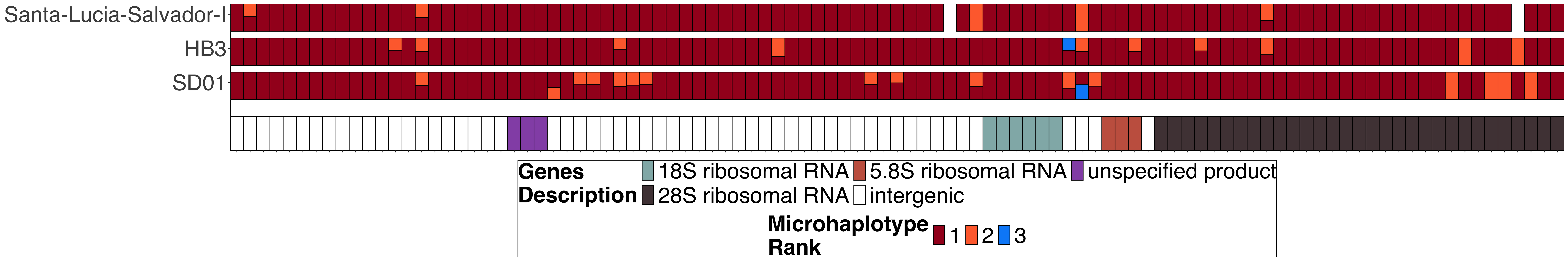
Outputting the regions within the shared region where SD01 has multiple variants
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sd01 = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates %>%
filter(s_Sample == "SD01") %>%
group_by(s_Sample, p_name) %>%
mutate(s_COI = length(unique(h_popUID)))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sd01_multi = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sd01 %>%
filter(s_COI > 1)
regions_withSD01MultiInShared = regions %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sd01_multi$p_name)
write_tsv(regions_withSD01MultiInShared, "regions_withSD01MultiInShared.bed")
finalHrpSubwindows_regions_withSD01MultiInShared = finalHrpSubwindows %>%
filter(X4 %in% regions_withSD01MultiInShared$name)
write_tsv(finalHrpSubwindows_regions_withSD01MultiInShared, "finalHrpSubwindows_regions_withSD01MultiInShared.bed", col_names = F)popClustering_labIso = popClustering %>%
left_join(meta %>%
rename(s_Sample = BiologicalSample)) %>%
filter(grepl("^Pf", sample))
popClustering_labIso_homologousRegion = popClustering_labIso %>%
filter(p_name %in% regions_homologousRegion$genomicID)
popClustering_labIso_homologousRegion_prep = HaplotypeRainbows::prepForRainbow(popClustering_labIso_homologousRegion, minPopSize = 1)# select just the major haplotypes and cluster based on the sharing between
popClustering_labIso_homologousRegion_prep_sp = popClustering_labIso_homologousRegion_prep %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample)))%>%
group_by()%>%
filter(sampleCount > 0.9*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_labIso_homologousRegion_prep_sp_mat = as.matrix(popClustering_labIso_homologousRegion_prep_sp[,2:ncol(popClustering_labIso_homologousRegion_prep_sp)])
rownames(popClustering_labIso_homologousRegion_prep_sp_mat) = popClustering_labIso_homologousRegion_prep_sp$s_Sample
popClustering_labIso_homologousRegion_prep_sp_dist = dist(popClustering_labIso_homologousRegion_prep_sp_mat)
popClustering_labIso_homologousRegion_prep_sp_dist_hclust = hclust(popClustering_labIso_homologousRegion_prep_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_labIso_homologousRegion_prep = popClustering_labIso_homologousRegion_prep %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_labIso_homologousRegion_prep_sp_mat)[popClustering_labIso_homologousRegion_prep_sp_dist_hclust$order]))
popClustering_labIso_homologousRegion_prep_plot = genRainbowHapPlotObj(popClustering_labIso_homologousRegion_prep, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_labIso_homologousRegion_prep$p_name)),
labels = levels(popClustering_labIso_homologousRegion_prep$p_name),
expand = c(0,0))+
scale_y_continuous(expand = c(0,0))The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal
regions_homologousRegion_filt = regions_homologousRegion %>%
filter(genomicID %in% popClustering_labIso_homologousRegion_prep$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_labIso_homologousRegion_prep$p_name)))
print(popClustering_labIso_homologousRegion_prep_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -1,
fill = description),
data = regions_homologousRegion_filt, color = "black") +
scale_fill_manual(values = descriptionColors_homologousRegion,
guide = guide_legend(nrow = 2))
)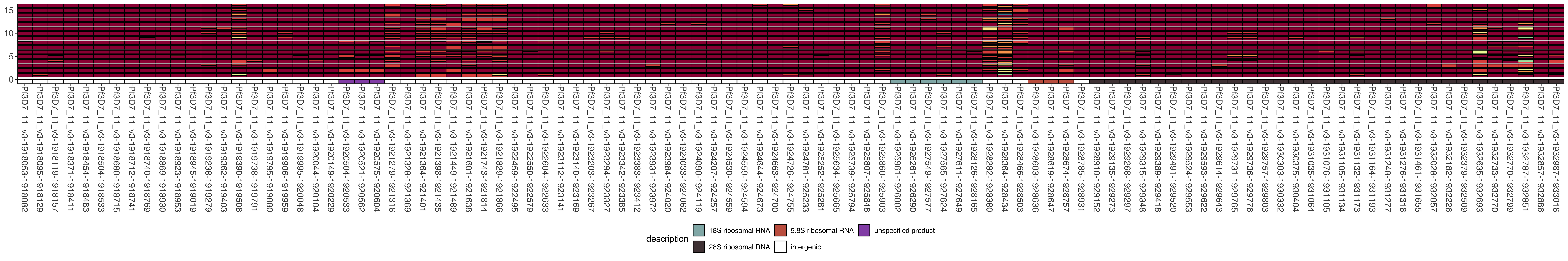
allSel = readr::read_tsv("/Users/nick/Dropbox (Personal)/ownCloud/documents/plasmodium/falciparum/pfepipanels/Pf_Epi_Panels/data/MIPSIBC/data/pf/reports/slim_allSelectedClustersInfo.tab.txt.gz")
allSel_filt = allSel %>%
filter(s_Sample %in% previousDeletionCalls$BiologicalSample)
write_tsv(allSel_filt, "MIPSIBC_previousDeletionCalls_samples.tsv")allSel = readr::read_tsv("/Users/nick/Dropbox (Personal)/ownCloud/documents/plasmodium/falciparum/pfepipanels/Pf_Epi_Panels/data/heome1/data/pf/reports/slim_allSelectedClustersInfo.tab.txt.gz")
allSel_filt = allSel %>%
filter(s_Sample %in% previousDeletionCalls$BiologicalSample)
write_tsv(allSel_filt, "heome1_previousDeletionCalls_samples.tsv")elucidator doPairwiseComparisonOnHapsSharing --tableFnp heome1_previousDeletionCalls_samples.tsv --sampleCol s_Sample --targetNameCol p_name --popIDCol h_popUID --relAbundCol c_AveragedFrac --numThreads 12 --dout heome1_previousDeletionCalls_samples_pairwiseComps --verbose --overWriteDir --metaFnp /tank/data/plasmodium/falciparum/pfdata/metadata/metaByBiosample.tab.txt --metaFieldsToCalcPopDiffs country,region,secondaryRegion --writeOutDistMatrices
elucidator doPairwiseComparisonOnHapsSharing --tableFnp MIPSIBC_previousDeletionCalls_samples.tsv --sampleCol s_Sample --targetNameCol p_name --popIDCol h_popUID --relAbundCol c_AveragedFrac --numThreads 12 --dout MIPSIBC_previousDeletionCalls_samples_pairwiseComps --verbose --overWriteDir --metaFnp /tank/data/plasmodium/falciparum/pfdata/metadata/metaByBiosample.tab.txt --metaFieldsToCalcPopDiffs country,region,secondaryRegion --writeOutDistMatrices sample_metadata_withAllDeletionCalls=readr::read_tsv("sample_metadata_withAllDeletionCalls.tsv")
heome1_previousDeletionCalls_samples_jacardByHapsTarShared_samples = readr::read_tsv("heome1_previousDeletionCalls_samples_pairwiseComps/sampleNames.tab.txt", col_names = F)
heome1_previousDeletionCalls_samples_jacardByHapsTarShared = readr::read_tsv("heome1_previousDeletionCalls_samples_pairwiseComps/jacardByHapsTarShared.tab.txt.gz", col_names = F)
heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat = as.matrix(heome1_previousDeletionCalls_samples_jacardByHapsTarShared)
colnames(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat) = heome1_previousDeletionCalls_samples_jacardByHapsTarShared_samples$X1
rownames(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat) = heome1_previousDeletionCalls_samples_jacardByHapsTarShared_samples$X1
heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat = heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat[metaSelected$BiologicalSample,metaSelected$BiologicalSample]
col_fun = colorRamp2(c(min(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat), min(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat) + (1-min(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
previousDeletionCalls_sel = previousDeletionCalls[match(colnames(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat), previousDeletionCalls$BiologicalSample),]%>%
left_join(sample_metadata_withAllDeletionCalls %>%
select(sample, Pattern) %>%
rename(BiologicalSample = sample)) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
mutate(newClusterName = ifelse(hcclustSize == 1, "singlet", as.character(stringr::str_pad(newClusterName, width = 2, pad = "0")))) %>%
mutate(BiologicalSample = as.character(BiologicalSample)) %>%
select(BiologicalSample, newClusterName))
rowAnnoDf = previousDeletionCalls_sel[, c(
"country",
"region",
"secondaryRegion",
"newClusterName",
"Pattern")] %>%
rename(continent = secondaryRegion,
Chr11DupHapCluster = newClusterName) %>%
as.data.frame()
rowAnnoColors = createColorListFromDf(rowAnnoDf)
load("rowAnnoColors.Rdata")
rowAnnoColors_mod = rowAnnoColors
rowAnnoColors_mod[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
rowAnnoColors_mod[["Pattern"]] = rowAnnoColorsMod_hrp3DeletionPattern
annotationTextSize = 25 ;annotationTitleTextSize = 20;
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
show_legend = F,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm = Heatmap(
heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat,
col = col_fun,
name = "JacardIndex",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
na_col = "#FFFFFF00",
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "JacardIndex"
)
)
)draw(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")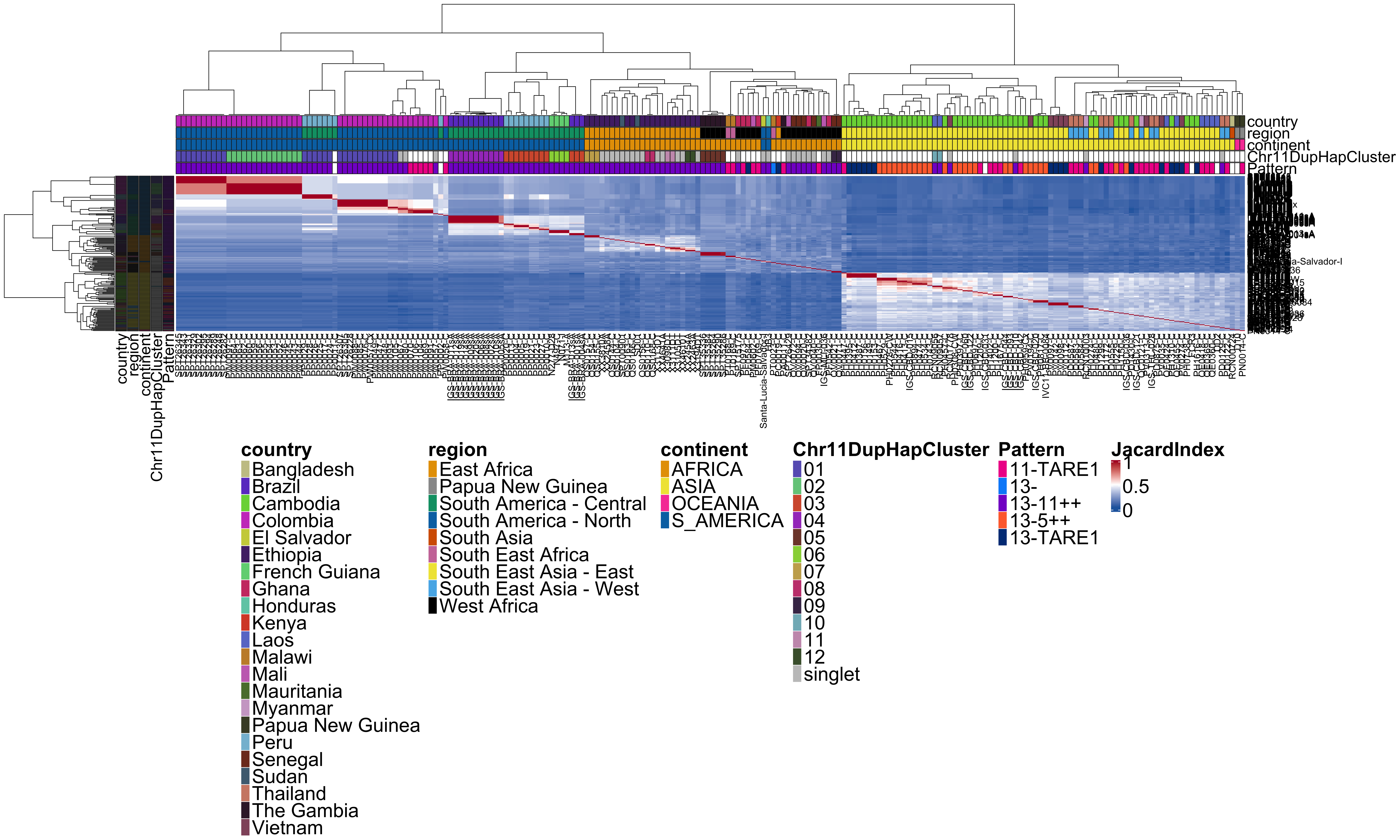
pdf("heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm.pdf", width = 25, height = 30, useDingbats = F)
draw(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
dev.off()quartz_off_screen
2 sample_metadata_withAllDeletionCalls=readr::read_tsv("sample_metadata_withAllDeletionCalls.tsv")
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_samples = readr::read_tsv("MIPSIBC_previousDeletionCalls_samples_pairwiseComps/sampleNames.tab.txt", col_names = F)
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared = readr::read_tsv("MIPSIBC_previousDeletionCalls_samples_pairwiseComps/jacardByHapsTarShared.tab.txt.gz", col_names = F)
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat = as.matrix(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared)
colnames(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat) = MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_samples$X1
rownames(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat) = MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_samples$X1
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat = MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat[metaSelected$BiologicalSample,metaSelected$BiologicalSample]
col_fun = colorRamp2(c(min(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat), min(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat) + (1-min(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
previousDeletionCalls_sel = previousDeletionCalls[match(colnames(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat), previousDeletionCalls$BiologicalSample),]%>%
left_join(sample_metadata_withAllDeletionCalls %>%
select(sample, Pattern) %>%
rename(BiologicalSample = sample)) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
mutate(newClusterName = ifelse(hcclustSize == 1, "singlet", as.character(stringr::str_pad(newClusterName, width = 2, pad = "0")))) %>%
mutate(BiologicalSample = as.character(BiologicalSample)) %>%
select(BiologicalSample, newClusterName))
rowAnnoDf = previousDeletionCalls_sel[, c(
"country",
"region",
"secondaryRegion",
"newClusterName",
"Pattern")] %>%
rename(continent = secondaryRegion,
Chr11DupHapCluster = newClusterName) %>%
as.data.frame()
rowAnnoColors = createColorListFromDf(rowAnnoDf)
load("rowAnnoColors.Rdata")
rowAnnoColors_mod = rowAnnoColors
rowAnnoColors_mod[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
rowAnnoColors_mod[["Pattern"]] = rowAnnoColorsMod_hrp3DeletionPattern
annotationTextSize = 25 ;annotationTitleTextSize = 20;
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
show_legend = F,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_nolabs = MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat
rownames(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_nolabs) = NULL
colnames(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_nolabs) = NULL
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm = Heatmap(
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_nolabs,
col = col_fun,
name = "JacardIndex",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
na_col = "#FFFFFF00",
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "JacardIndex"
)
)
)draw(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")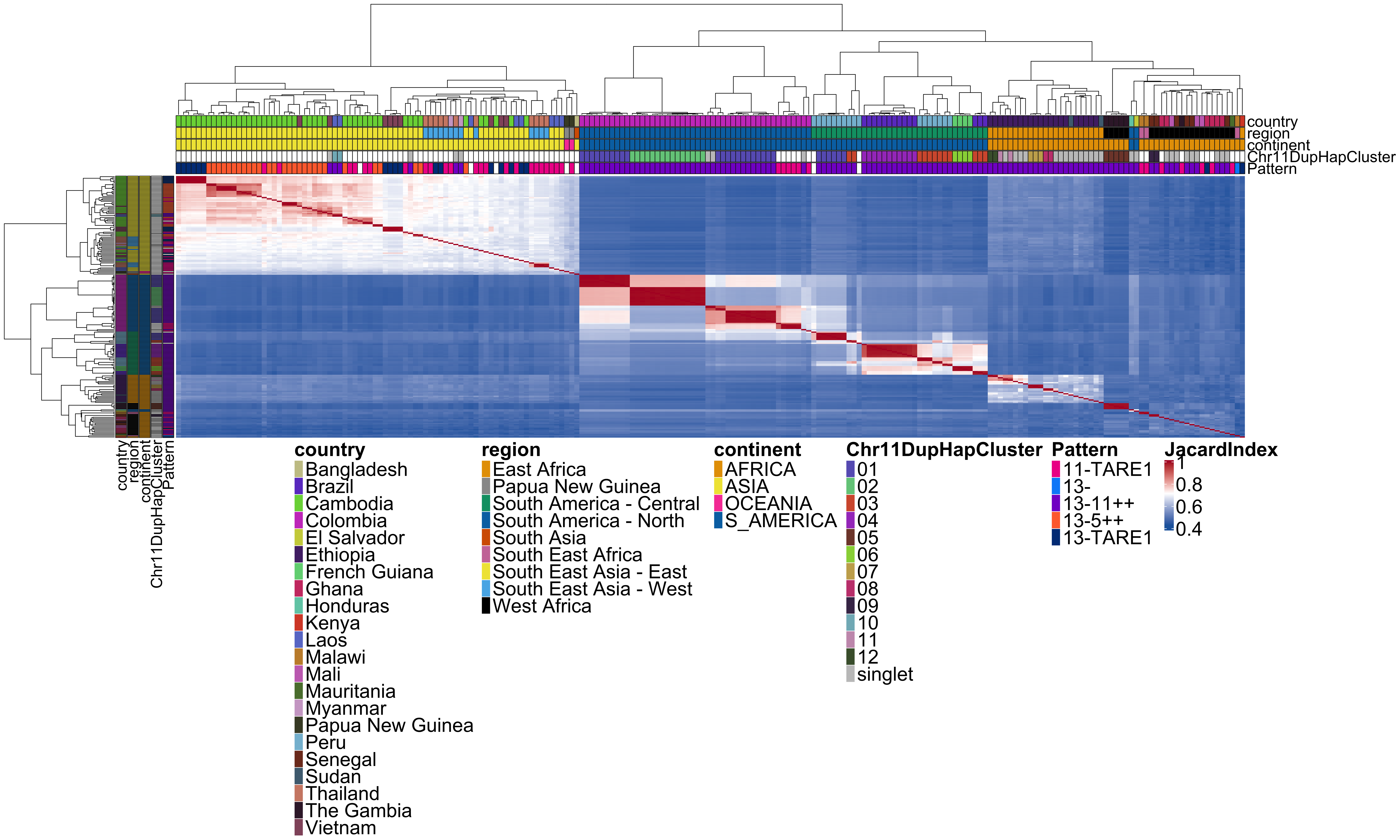
pdf("MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm.pdf", width = 25, height = 30, useDingbats = F)
draw(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
dev.off()quartz_off_screen
2 sample_metadata_withAllDeletionCalls_sel = sample_metadata_withAllDeletionCalls %>%
select(sample, SRARuns) %>%
filter(sample %in% previousDeletionCalls$BiologicalSample) %>%
mutate(SRARuns = strsplit(SRARuns, split = ",")) %>%
unnest(SRARuns)
hmm_fract = readr::read_tsv("filtered_gatk_calls_database_hrpsallMetaDeletionCalls.hmm_fract.txt")
hmm_fract_combined = bind_rows(
hmm_fract %>%
arrange(sample1, sample2) %>%
select(sample1, sample2, fract_sites_IBD),
hmm_fract %>%
arrange(sample1, sample2) %>%
select(sample1, sample2, fract_sites_IBD) %>%
rename(temp1 = sample2,
temp2 = sample1)%>%
rename(sample1 = temp1,
sample2 = temp2)
)
hmm_fract_combined_samples = tibble(sample1 = unique(hmm_fract_combined$sample1)) %>%
left_join(sample_metadata_withAllDeletionCalls_sel %>%
rename(BiologicalSample = sample,
sample1 = SRARuns)) %>%
mutate(BiologicalSample = ifelse(is.na(BiologicalSample), sample1, BiologicalSample)) %>%
mutate(BiologicalSample = ifelse(BiologicalSample == "fcr3", "FCR3", BiologicalSample))
perSampleVarCounts = readr::read_tsv("filtered_hrpsallMetaDeletionCalls_variants_perSampleCounts.tsv") %>%
rename(sample1 = `[3]sample`,
nMissing = `[14]nMissing`)
hmm_fract_combined_samples_filt = hmm_fract_combined_samples %>%
left_join(perSampleVarCounts %>%
select(sample1, nMissing)) %>%
group_by(BiologicalSample) %>%
arrange(BiologicalSample, nMissing) %>%
mutate(rank = row_number(),
totalSamples = n()) %>%
filter(rank == 1)
hmm_fract_combined_filt = hmm_fract_combined %>%
filter(sample1 %in% hmm_fract_combined_samples_filt$sample1,
sample2 %in% hmm_fract_combined_samples_filt$sample1) %>%
left_join(hmm_fract_combined_samples_filt %>%
rename(BiologicalSample1 = BiologicalSample) %>%
select(sample1, BiologicalSample1))%>%
left_join(hmm_fract_combined_samples_filt %>%
rename(
sample2 = sample1,
BiologicalSample2 = BiologicalSample) %>%
select(sample2, BiologicalSample2))
hmm_fract_sp = hmm_fract_combined_filt %>%
select(BiologicalSample1, BiologicalSample2, fract_sites_IBD) %>%
spread(BiologicalSample2, fract_sites_IBD, fill = 1)
hmm_fract_sp_mat = as.matrix(hmm_fract_sp[,2:ncol(hmm_fract_sp)])
rownames(hmm_fract_sp_mat) = hmm_fract_sp$BiologicalSample1library(circlize)
hmm_fract_sp_mat = hmm_fract_sp_mat[metaSelected$BiologicalSample, metaSelected$BiologicalSample]
# col_fun = colorRamp2(c(min(hmm_fract_sp_mat), min(hmm_fract_sp_mat) + (1-min(hmm_fract_sp_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
col_fun = colorRamp2(c(0, 0.5, 1), c( "#2166ac", "white", "#b2182b"))
previousDeletionCalls_sel = previousDeletionCalls[match(colnames(hmm_fract_sp_mat), previousDeletionCalls$BiologicalSample),]%>%
left_join(sample_metadata_withAllDeletionCalls %>%
select(sample, Pattern) %>%
rename(BiologicalSample = sample)) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
mutate(newClusterName = ifelse(hcclustSize == 1, "singlet", as.character(stringr::str_pad(newClusterName, width = 2, pad = "0")))) %>%
mutate(BiologicalSample = as.character(BiologicalSample)) %>%
select(BiologicalSample, newClusterName)) %>%
mutate(Pattern = ifelse(is.na(Pattern) & possiblyHRP2Deleted, "8-TARE1", Pattern)) %>%
mutate(Pattern = ifelse("FCR3" == BiologicalSample, "13++11-", Pattern))
rowAnnoDf = previousDeletionCalls_sel[, c(
"country",
"region",
"secondaryRegion",
"newClusterName",
"Pattern",
"hrpCall")] %>%
rename(continent = secondaryRegion,
Chr11DupHapCluster = newClusterName) %>%
as.data.frame()
rowAnnoColors = createColorListFromDf(rowAnnoDf)
load("rowAnnoColors.Rdata")
rowAnnoColors_mod = rowAnnoColors
rowAnnoColors_mod[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
rowAnnoColors_mod[["Pattern"]] = rowAnnoColorsMod_hrp3DeletionPattern
annotationTextSize = 25 ;annotationTitleTextSize = 20;
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
show_legend = F,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
hmm_fract_sp_mat_nolabs = hmm_fract_sp_mat
rownames(hmm_fract_sp_mat_nolabs) = NULL
colnames(hmm_fract_sp_mat_nolabs) = NULL
hmm_fract_sp_mat_hm = Heatmap(
hmm_fract_sp_mat_nolabs,
col = col_fun,
name = "fracIBDSites",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
na_col = "#FFFFFF00" ,
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "fracIBDSites"
)
)
)draw(hmm_fract_sp_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
quartz_off_screen
2 hmm_fract_sp_pat1 = hmm_fract_combined_filt %>%
filter(BiologicalSample1 %in% metaSelected_hrp3_pat1$BiologicalSample,
BiologicalSample2 %in% metaSelected_hrp3_pat1$BiologicalSample) %>%
select(BiologicalSample1, BiologicalSample2, fract_sites_IBD) %>%
spread(BiologicalSample2, fract_sites_IBD, fill = 1)
hmm_fract_sp_pat1_mat = as.matrix(hmm_fract_sp_pat1[,2:ncol(hmm_fract_sp_pat1)])
rownames(hmm_fract_sp_pat1_mat) = hmm_fract_sp_pat1$BiologicalSample1library(circlize)
# col_fun = colorRamp2(c(min(hmm_fract_sp_pat1_mat), min(hmm_fract_sp_pat1_mat) + (1-min(hmm_fract_sp_pat1_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
col_fun = colorRamp2(c(0, 0.5, 1), c( "#2166ac", "white", "#b2182b"))
previousDeletionCalls_sel = previousDeletionCalls[match(colnames(hmm_fract_sp_pat1_mat), previousDeletionCalls$BiologicalSample),]%>%
left_join(sample_metadata_withAllDeletionCalls %>%
select(sample, Pattern) %>%
rename(BiologicalSample = sample)) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
mutate(newClusterName = ifelse(hcclustSize == 1, "singlet", as.character(stringr::str_pad(newClusterName, width = 2, pad = "0")))) %>%
mutate(BiologicalSample = as.character(BiologicalSample)) %>%
select(BiologicalSample, newClusterName))
rowAnnoDf = previousDeletionCalls_sel[, c(
"country",
"region",
"secondaryRegion",
"newClusterName",
# "Pattern",
"hrpCall")] %>%
rename(continent = secondaryRegion,
Chr11DupHapCluster = newClusterName) %>%
as.data.frame()
rowAnnoColors = createColorListFromDf(rowAnnoDf)
load("rowAnnoColors.Rdata")
rowAnnoColors_mod = rowAnnoColors
rowAnnoColors_mod[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
rowAnnoColors_mod[["Pattern"]] = rowAnnoColorsMod_hrp3DeletionPattern
annotationTextSize = 25 ;annotationTitleTextSize = 20;
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
show_legend = F,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
hmm_fract_sp_pat1_mat_nolabs = hmm_fract_sp_pat1_mat
rownames(hmm_fract_sp_pat1_mat_nolabs) = NULL
colnames(hmm_fract_sp_pat1_mat_nolabs) = NULL
hmm_fract_sp_pat1_mat_hm = Heatmap(
hmm_fract_sp_pat1_mat_nolabs,
col = col_fun,
name = "fracIBDSites",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
na_col = "#FFFFFF00" ,
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "fracIBDSites"
)
)
)draw(hmm_fract_sp_pat1_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
sample_metadata_withAllDeletionCalls_13_5_pattern = sample_metadata_withAllDeletionCalls %>%
filter(Pattern == "13-5++")
hmm_fract_sp_13_5_pattern = hmm_fract_combined_filt %>%
filter(BiologicalSample1 %in% sample_metadata_withAllDeletionCalls_13_5_pattern$sample,
BiologicalSample2 %in% sample_metadata_withAllDeletionCalls_13_5_pattern$sample) %>%
select(BiologicalSample1, BiologicalSample2, fract_sites_IBD) %>%
spread(BiologicalSample2, fract_sites_IBD, fill = 1)
hmm_fract_sp_13_5_pattern_mat = as.matrix(hmm_fract_sp_13_5_pattern[,2:ncol(hmm_fract_sp_13_5_pattern)])
rownames(hmm_fract_sp_13_5_pattern_mat) = hmm_fract_sp_13_5_pattern$BiologicalSample1library(circlize)
# col_fun = colorRamp2(c(min(hmm_fract_sp_13_5_pattern_mat), min(hmm_fract_sp_13_5_pattern_mat) + (1-min(hmm_fract_sp_13_5_pattern_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
col_fun = colorRamp2(c(0, 0.5, 1), c( "#2166ac", "white", "#b2182b"))
previousDeletionCalls_sel = previousDeletionCalls[match(colnames(hmm_fract_sp_13_5_pattern_mat), previousDeletionCalls$BiologicalSample),]%>%
left_join(sample_metadata_withAllDeletionCalls %>%
select(sample, Pattern) %>%
rename(BiologicalSample = sample)) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
mutate(newClusterName = ifelse(hcclustSize == 1, "singlet", as.character(stringr::str_pad(newClusterName, width = 2, pad = "0")))) %>%
mutate(BiologicalSample = as.character(BiologicalSample)) %>%
select(BiologicalSample, newClusterName))
rowAnnoDf = previousDeletionCalls_sel[, c(
"country",
"region",
"secondaryRegion"
# "newClusterName",
# "Pattern",
# "hrpCall"
)] %>%
rename(continent = secondaryRegion) %>%
as.data.frame()
rowAnnoColors = createColorListFromDf(rowAnnoDf)
load("rowAnnoColors.Rdata")
rowAnnoColors_mod = rowAnnoColors
# rowAnnoColors_mod[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
rowAnnoColors_mod[["Pattern"]] = rowAnnoColorsMod_hrp3DeletionPattern
annotationTextSize = 25 ;annotationTitleTextSize = 20;
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
show_legend = F,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
hmm_fract_sp_13_5_pattern_mat_nolabs = hmm_fract_sp_13_5_pattern_mat
rownames(hmm_fract_sp_13_5_pattern_mat_nolabs) = NULL
colnames(hmm_fract_sp_13_5_pattern_mat_nolabs) = NULL
hmm_fract_sp_13_5_pattern_mat_hm = Heatmap(
hmm_fract_sp_13_5_pattern_mat_nolabs,
col = col_fun,
name = "fracIBDSites",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
na_col = "#FFFFFF00" ,
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "fracIBDSites"
)
)
)draw(hmm_fract_sp_13_5_pattern_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")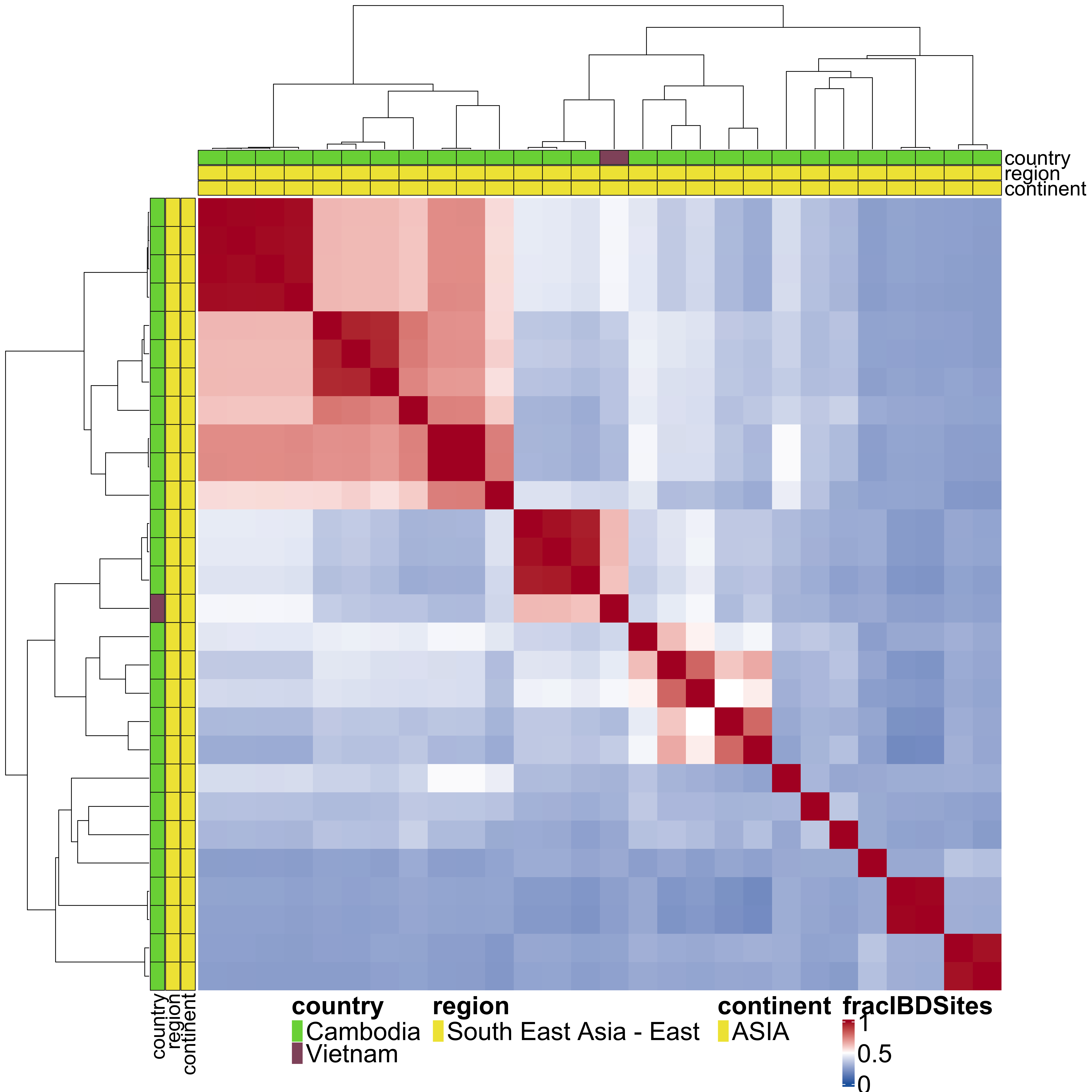
remotes::install_github("EPPIcenter/moire")
setwd("/tank/data/plasmodium/falciparum/pfdata/moire_on_hrp3Samples/")
df <- read.tsv("allSel_withDeletions_prep_outForMoire.tsv")
data <- load_long_form_data(df)
# With data in appropriate format, run MCMC as follows
mcmc_results <- moire::run_mcmc(data, is_missing = data$is_missing)
write_rds(mcmc_results, "mcmc_results.rds")
write_rds(data, "data_for_moire.rds")data_for_moire = read_rds("moire_on_hrp3Samples/data_for_moire.rds")
mcmc_results = read_rds("moire_on_hrp3Samples/mcmc_results.rds")
coiEsts = tibble(
sampleID = data_for_moire$sample_ids,
medianCOI = round(unlist(lapply(mcmc_results$chains[[1]]$coi, median))),
meanCOI = round(unlist(lapply(mcmc_results$chains[[1]]$coi, mean))),
maxCOI = round(unlist(lapply(mcmc_results$chains[[1]]$coi, max)))
)---
title: "Plotting haplotype variation within regions"
---
```{r setup, echo=FALSE, message=FALSE}
source("../common.R")
```
## Reading in data
```{r}
load("rowAnnoColors.Rdata")
```
## Downloads
```{r}
#| results: asis
#| echo: false
cat(createDownloadLink("../meta/metadata/meta.tab.txt"))
cat(createDownloadLink("../meta/metadata/metaByBioSample"))
cat(createDownloadLink("wgs_variants/THEREALMcCOIL/categorical_method/real_mccoil_COI_calls.tsv"))
cat(createDownloadLink("finalHRPII_HRPIII_windows_withTunedSubWindows/popClustering/reports/slim_allSelectedClustersInfo.tab.txt.gz"))
cat(createDownloadLink("metaSelected.tab.txt"))
cat(createDownloadLink("allMeta_HRP2_HRP3_deletionCalls.tab.txt"))
cat(createDownloadLink("subwindows_regionMeta.tab.txt"))
```
```{r}
meta = readr::read_tsv("../meta/metadata/meta.tab.txt") %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
metaByBioSample = readr::read_tsv("../meta/metadata/metaByBioSample.tab.txt") %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
# coiCalls = readr::read_tsv("/Users/nick/Dropbox (Personal)/ownCloud/documents/plasmodium/falciparum/pfepipanels/Pf_Epi_Panels/data/MAD4HATTER/data/pf/COI_calls.tab.txt")
# coiCalls_poly = coiCalls %>%
# filter(COI > 1)
coiCalls = readr::read_tsv("heome1_COI_calls.tab.txt")
#coiCalls = readr::read_tsv("PfSMART_COI_calls.tab.txt")
coiCalls_poly = coiCalls %>%
filter(COI > 1)
realmccoilCoiCalls = readr::read_tsv("wgs_variants/THEREALMcCOIL/categorical_method/real_mccoil_COI_calls.tsv")
realmccoilCoiCalls_poly = realmccoilCoiCalls %>%
filter(random_median != 1 | topHE_median != 1)
previousDeletionCalls = readr::read_tsv("allMeta_HRP2_HRP3_deletionCalls.tab.txt") %>%
#filter(country %!in% c("Bangladesh", "Mauritania", "Myanmar", "The Gambia")) %>%
#filter(((grepl("SPT", sample) & possiblyChr11Deleted))) %>%
#filter(BiologicalSample %!in% coiCalls_poly$sample) %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
meta = meta %>%
left_join(previousDeletionCalls)%>%
mutate(hrpCall = case_when(
possiblyHRP2Deleted & possiblyHRP3Deleted ~ "pfhrp2-/pfhrp3-",
possiblyHRP2Deleted & !possiblyHRP3Deleted ~ "pfhrp2-/pfhrp3+",
!possiblyHRP2Deleted & possiblyHRP3Deleted ~ "pfhrp2+/pfhrp3-",
T ~ "pfhrp2+/pfhrp3+"
)) %>%
left_join(realmccoilCoiCalls %>%
select(BiologicalSample, topHE_median) %>%
rename(COI = topHE_median))
# left_join(coiCalls %>%
# rename(BiologicalSample = sample))
homologousRegion = readr::read_tsv("../rRNA_segmental_duplications/sharedBetween11_and_13/investigatingChrom11Chrom13/Pf3D7_13_v3-2792021-2807295-for--Pf3D7_11_v3-1918028-1933288-for.bed",
col_names = F)
regions = readr::read_tsv("subwindows_regionMeta.tab.txt")
metaSelected = readr::read_tsv("metaSelected.tab.txt") %>%
#select(-COI) %>%
left_join(realmccoilCoiCalls %>%
select(BiologicalSample, topHE_median) %>%
rename(COI = topHE_median)) %>%
filter(COI == 1)
metaSelected_hrp2_deleted = metaSelected %>% filter(possiblyHRP2Deleted)
metaSelected_hrp3_deleted = metaSelected %>% filter(possiblyHRP3Deleted)
metaSelected_hrp2_and_hrp3_deleted = metaSelected %>% filter(possiblyHRP2Deleted, possiblyHRP3Deleted)
regions_key = regions %>%
select(name, genomicID)
```
```{r}
finalHrpSubwindows = readr::read_tsv("../windowAnalysis/windows/finalHRPII_HRPIII_windows_withTuned_combinedVarConservedRegions.bed", col_names = F)
erroneousRegions = c("Pf3D7_11_v3-1944071-1944237", "Pf3D7_11_v3-1944083-1944229", "Pf3D7_11_v3-1938175-1938354")
samplesCovered = readr::read_tsv("samplesCovered.txt", col_names = "sample") %>%
left_join(meta %>%
select(sample, BiologicalSample))
```
```{r}
popClustering = readr::read_tsv("finalHRPII_HRPIII_windows_withTunedSubWindows/popClustering/reports/slim_allSelectedClustersInfo.tab.txt.gz")
#
regions_key = regions_key %>%
mutate(duplicationRegion = grepl("for", name))
# renaming and duplicate the dup region
popClustering = popClustering %>%
left_join(regions_key %>%
rename(p_name = name)) %>%
mutate(p_name = genomicID) %>%
mutate(h_popUID = paste0(genomicID, "--", h_popUID))
#
popClustering_filt = popClustering %>%
filter(s_Sample %fin% metaSelected$BiologicalSample) %>%
filter(genomicID %!in% erroneousRegions)
previousDeletionCalls_hrp3Delete = previousDeletionCalls %>%
filter(possiblyHRP3Deleted)
popClustering_filt_hrp3Delete = popClustering_filt %>%
filter(s_Sample %in% previousDeletionCalls_hrp3Delete$BiologicalSample)
regions_afterHomologous = regions %>%
filter(afterHomologousRegion)
```
```{r}
allDeletionTypeMeta = readr::read_tsv("allMetaDeletionCalls.tab.txt") %>%
filter(BiologicalSample %in% metaSelected$BiologicalSample)
allDeletionTypeMeta_hrp3_pat1 = allDeletionTypeMeta %>%
filter(HRP3_deletionPattern == "Pattern 1")
popClustering_filt_hrp3_pat1 = popClustering_filt %>%
filter(s_Sample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample)
allDeletionTypeMeta_hrp3_pat2 = allDeletionTypeMeta %>%
filter(HRP3_deletionPattern == "Pattern 2")
popClustering_filt_hrp3_pat2 = popClustering_filt %>%
filter(s_Sample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample)
allDeletionTypeMeta_deletionPatternCounts = allDeletionTypeMeta %>%
filter(!is.na(HRP3_deletionPattern)) %>%
group_by(HRP3_deletionPattern) %>%
count()
create_dt(allDeletionTypeMeta_deletionPatternCounts)
```
## Pattern 2
```{r}
allDeletionTypeMeta_hrp3_pat2_count_country = allDeletionTypeMeta_hrp3_pat2 %>%
group_by(country, region, secondaryRegion) %>%
count()
create_dt(allDeletionTypeMeta_hrp3_pat2_count_country)
allDeletionTypeMeta_hrp3_pat2_count_region = allDeletionTypeMeta_hrp3_pat2 %>%
group_by(region, secondaryRegion) %>%
count()
create_dt(allDeletionTypeMeta_hrp3_pat2_count_region)
allDeletionTypeMeta_hrp3_pat2_count_continent = allDeletionTypeMeta_hrp3_pat2 %>%
group_by(secondaryRegion) %>%
count()
create_dt(allDeletionTypeMeta_hrp3_pat2_count_continent)
```
## 13-11++
### Chr 11 duplicated region
#### Getting chr 11 duplication conserved counts
Below is code determining the samples with possible chr11 fragment duplication and breaking down the counts of perfect duplicated copies vs divergent copies.
```{r}
regions_afterHomologous_chr11 = regions %>%
filter(`#chrom` == "Pf3D7_11_v3",
afterHomologousRegion)
regions_afterHomologous_chr11 = regions_afterHomologous_chr11 %>%
mutate(description = case_when(
grepl("extraField0=NA", extraField0) ~ "intergenic",
T ~ gsub("\\]", "", gsub(".*description=", "", extraField0))
) )
descriptionColors = scheme$hex(length(regions_afterHomologous_chr11$description %>% unique()))
names(descriptionColors) = regions_afterHomologous_chr11$description %>% unique()
descriptionColors["intergenic"] = c("#FF000000")
```
```{r}
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 = popClustering_filt_hrp3_pat1 %>%
filter(p_name %in% regions_afterHomologous_chr11$genomicID)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
group_by(s_Sample, p_name) %>%
mutate(uniqHaps= n())
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_uniqSum = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
group_by(s_Sample) %>%
mutate(targets = length(unique(genomicID))) %>%
group_by(s_Sample, targets, uniqHaps) %>%
count() %>%
mutate(freq = n/targets)
minafCutoff = 0.15
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
filter(c_AveragedFrac > minafCutoff) %>%
group_by(s_Sample, p_name) %>%
mutate(uniqHaps= n()) %>%
mutate(marker = uniqHaps == 1) %>%
group_by(s_Sample) %>%
summarise(conserved = sum(marker),
targets = length(unique(genomicID))) %>%
mutate(conservedID = conserved/targets)
conservedCutOff = 0.99
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum %>%
filter(conservedID > conservedCutOff)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOff = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
group_by() %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOffByCountry = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
left_join(metaByBioSample %>%
rename(s_Sample = sample)) %>%
group_by(country, region) %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOffByRegion = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
left_join(metaByBioSample %>%
rename(s_Sample = sample)) %>%
group_by(region) %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOffByContinent = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
left_join(metaByBioSample %>%
rename(s_Sample = sample)) %>%
group_by(secondaryRegion) %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)
```
The number of samples with perfect copies
```{r}
create_dt(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOff)
```
The number of samples with perfect copies broken down by country
```{r}
create_dt(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOffByCountry)
```
The number of samples with perfect copies broken down by regions
```{r}
create_dt(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOffByRegion)
```
The number of samples with perfect copies broken down by continent.
```{r}
create_dt(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_cutOffByContinent)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_meanId = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum %>%
filter(conservedID <= conservedCutOff) %>%
summarise(meanID = mean(conservedID),
minID = min(conservedID),
sdID = sd(conservedID))
```
The breakdown of level of divergence in the samples with divergent samples.
```{r}
create_dt(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_meanId)
```
## Population analysis of chr11 duplicated region
Calculating the population of the haplotypes after the shared region on chr 11, the duplicated region to see if there is any population signal associated with the duplicated copy. E.g. if the copy is unique to a subset of haplotypes, if the copy is always perfect or if there is variation.
```{r}
popClustering_filt_regions_afterHomologous_chr11 = popClustering %>%
filter(genomicID %!in% erroneousRegions) %>%
filter(p_name %in% regions_afterHomologous_chr11$genomicID)
popClustering_filt_regions_afterHomologous_chr11_tarCounts = popClustering_filt_regions_afterHomologous_chr11 %>%
group_by(s_Sample) %>%
summarise(tarCounts = length(unique(p_name)))
popClustering_filt_regions_afterHomologous_chr11_tarCounts_filt = popClustering_filt_regions_afterHomologous_chr11_tarCounts %>%
filter(tarCounts >= 0.80 * max(tarCounts) |
s_Sample %in% previousDeletionCalls$BiologicalSample)
popClustering_filt_regions_afterHomologous_chr11_sampCounts = popClustering_filt_regions_afterHomologous_chr11 %>%
group_by(p_name) %>%
summarise(sampCounts = length(unique(s_Sample)))
metaByBioSample_out = metaByBioSample %>%
left_join(allDeletionTypeMeta %>%
select(-sample, -ExperimentSample) %>%
rename(sample = BiologicalSample))
write_tsv(metaByBioSample_out, "metaByBioSample_outwithHrpCalls.tab.txt")
write_tsv(popClustering_filt_regions_afterHomologous_chr11 %>%
filter(s_Sample %in% popClustering_filt_regions_afterHomologous_chr11_tarCounts_filt$s_Sample) %>%
group_by() %>%
select(s_Sample, p_name, h_popUID, c_AveragedFrac),
"popClustering_filt_regions_afterHomologous_chr11.tab.txt.gz")
```
```{bash, eval = F}
elucidator doPairwiseComparisonOnHapsSharingDev --tableFnp popClustering_filt_regions_afterHomologous_chr11.tab.txt.gz --sampleCol s_Sample --targetNameCol p_name --popIDCol h_popUID --relAbundCol c_AveragedFrac --numThreads 14 --dout pairwiseComps_regions_afterHomologous_chr11 --verbose --overWriteDir --metaFnp metaByBioSample_outwithHrpCalls.tab.txt --metaFieldsToCalcPopDiffs country,region,secondaryRegion,HRP3_deletionPattern --writeOutDistMatrices
```
```{r}
#jacardDist = readr::read_tsv("pairwiseComps_regions_afterHomologous_chr11/percOfTarSharingAtLeastOneHap.tab.txt.gz", col_names = F)
jacardDist = readr::read_tsv("pairwiseComps_regions_afterHomologous_chr11/jacardByHapsTarShared.tab.txt.gz", col_names = F)
jacardDistSamps = readr::read_tsv("pairwiseComps_regions_afterHomologous_chr11/sampleNames.tab.txt", col_names = "samples")
colnames(jacardDist) = jacardDistSamps$samples
jacardDist$sample = jacardDistSamps$samples
# jacardDist_filt = jacardDist[jacardDist$sample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample,c(allDeletionTypeMeta_hrp3_pat1$BiologicalSample, "sample")]
# jacardDist_gat = jacardDist_filt %>%
# gather(otherSample, index,1:(ncol(.) - 1))
jacardDist_gat = jacardDist %>%
gather(otherSample, index,1:(ncol(.) - 1))
jacardDist_gat_filt = jacardDist_gat %>%
filter(sample %fin% allDeletionTypeMeta_hrp3_pat1$BiologicalSample,
otherSample %fin% allDeletionTypeMeta_hrp3_pat1$BiologicalSample)
jacardDist_gat_filt_sp = jacardDist_gat_filt %>%
spread(otherSample, index)
jacardDist_gat_filt_sp_mat = as.matrix(jacardDist_gat_filt_sp[,2:ncol(jacardDist_gat_filt_sp)])
rownames(jacardDist_gat_filt_sp_mat) = jacardDist_gat_filt_sp$sample
```
#### Getting cluster groups
```{r}
jacardDist_gat_filt_sp_mat_pat1 = jacardDist_gat_filt_sp_mat
# get data just for variable regions (e.g., minPopSize = 2)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11, minPopSize = 2)
# cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample))) %>%
group_by() %>%
filter(sampleCount >= 0.99*max(sampleCount)) %>% # get just targets with high sample coverage otherwise clustering will be by missingness
group_by(s_Sample, p_name) %>%
#filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist)
# get clustering based on the jacard distance too for reference
jacardDist_gat_filt_sp_mat_pat1_hc = hclust(dist(jacardDist_gat_filt_sp_mat_pat1))
k_groups = 42;
h_groups = 1.1;
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups = cutree(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust, k = k_groups)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend <- as.dendrogram(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend <- color_labels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend, k = k_groups)
plot(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups = cutree(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust, h = h_groups)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend <- as.dendrogram(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend <- color_labels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend, h = h_groups)
plot(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend)
pdf("popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend.pdf", height = 10, width = 20, useDingbats = F)
plot(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_dend)
dev.off()
jacardDist_gat_filt_sp_mat_pat1_hc_groups = cutree(jacardDist_gat_filt_sp_mat_pat1_hc, k = k_groups)
jacardDist_gat_filt_sp_mat_pat1_hc_dend <- as.dendrogram(jacardDist_gat_filt_sp_mat_pat1_hc)
jacardDist_gat_filt_sp_mat_pat1_hc_dend <- color_labels(jacardDist_gat_filt_sp_mat_pat1_hc_dend, k = k_groups)
plot(jacardDist_gat_filt_sp_mat_pat1_hc_dend)
pdf("jacardDist_gat_filt_sp_mat_pat1_hc_dend.pdf", height = 10, width = 20, useDingbats = F)
plot(jacardDist_gat_filt_sp_mat_pat1_hc_dend)
dev.off()
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df = tibble(
BiologicalSample = names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups),
hcclust = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups
) %>%
# mutate(BiologicalSample =factor(BiologicalSample, levels = levels(meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering$BiologicalSample))) %>%
group_by(hcclust) %>%
mutate(hcclustSize = n())
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_biggerGroups = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>% filter(hcclustSize != 1) %>% select(hcclust) %>% unique()
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_singletonGroups = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>% filter(hcclustSize == 1) %>% select(hcclust) %>% unique()
newscheme <- iwanthue(seed = 626, force_init = TRUE); newscheme$hex(8)
# nonSingletonGroupsColors = createColorListFromDf(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_biggerGroups)$hcclust
nonSingletonGroupsColors = newscheme$hex(nrow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_biggerGroups))
names(nonSingletonGroupsColors) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_biggerGroups$hcclust
nonSingletonGroupsColors_singleton = rep("grey71", nrow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_singletonGroups))
names(nonSingletonGroupsColors_singleton) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_singletonGroups$hcclust
haploGroupColors = c(nonSingletonGroupsColors, nonSingletonGroupsColors_singleton)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
select(hcclust, hcclustSize) %>%
ungroup() %>%
unique() %>%
arrange(desc(hcclustSize)) %>%
mutate(hcclust = as.character(hcclust),
newClusterName = row_number()) %>%
left_join(tibble(
hcclust = names(haploGroupColors),
colors = unname(haploGroupColors)
)) %>%
mutate(Chr11DupHapCluster = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, pad = "0", width = 2)))
newHaploGroupColors = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts$colors
names(newHaploGroupColors)= popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts$newClusterName
newHaploGroupWithSingletColors = c(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts$colors[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts$hcclustSize > 1],
"grey77")
names(newHaploGroupWithSingletColors)= c(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts$Chr11DupHapCluster[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts$hcclustSize > 1],
"singlet")
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_groupCounts %>%
mutate(hcclust = as.integer(hcclust)))
write_tsv(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df, "popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df.tsv")
```
```{r}
library(circlize)
#col_fun = colorRamp2(c(0, 0.5, 1), c(heat.colors(3)))
col_fun = colorRamp2(c(min(jacardDist_gat_filt_sp_mat), min(jacardDist_gat_filt_sp_mat) + (1-min(jacardDist_gat_filt_sp_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
jacardDist_gat_filt_sp_mat_noLabs = jacardDist_gat_filt_sp_mat
jacardDist_gat_filt_sp_mat_pat1 = jacardDist_gat_filt_sp_mat
meta_preferredSample = metaSelected %>%
filter(PreferredSample)
metaSelected_hrp3_pat1 = meta_preferredSample[match(rownames(jacardDist_gat_filt_sp_mat), meta_preferredSample$BiologicalSample), ]%>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
select(BiologicalSample, Chr11DupHapCluster))
metaSelected_hrp3_pat1 = metaSelected_hrp3_pat1 %>%
mutate(PerfectChr11Copy = BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies$s_Sample)
rownames(jacardDist_gat_filt_sp_mat_noLabs) = NULL
colnames(jacardDist_gat_filt_sp_mat_noLabs) = NULL
RowLabs = metaSelected_hrp3_pat1$BiologicalSample
RowLabs[metaSelected_hrp3_pat1$site != "LabIsolate" | is.na(metaSelected_hrp3_pat1$site)] = ""
ColLabs = metaSelected_hrp3_pat1$BiologicalSample
ColLabs[metaSelected_hrp3_pat1$site != "LabIsolate" | is.na(metaSelected_hrp3_pat1$site)] = ""
#RowLabs[metaSelected$country != "Ethiopia"] = ""
rownames(jacardDist_gat_filt_sp_mat_noLabs) = RowLabs
colnames(jacardDist_gat_filt_sp_mat_noLabs) = ColLabs
rowAnnoDf = metaSelected_hrp3_pat1[,c("hrpCall", "PerfectChr11Copy", "country", "region", "secondaryRegion", "Chr11DupHapCluster")] %>% rename(continent = secondaryRegion) %>% as.data.frame()
annotationTextSize = 25 ;annotationTitleTextSize = 20;
rowAnnoColors[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors,
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
show_legend = F,
gp = gpar(col = "grey10")
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
col = rowAnnoColors,
gp = gpar(col = "grey10")
)
haptype_hrp3_pat1HeatMap = Heatmap(
jacardDist_gat_filt_sp_mat_noLabs,
cluster_columns = T,
col = col_fun,
name = "JacardIndex",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "JacardIndex"
)
)
)
```
### Samples with a duplicated chromosome 11 and deleted chr 13 (13-11++ of HRP3 deletion)
Jacard index of the duplicated region on chromosome 11, jacard of 1 means complete agreement between samples on this region which 0 would be no haplotypes shared in this region. Additional meta data of the samples is shown on top and to the right including country/region, and the hrp2/3 calls, whether the the Chr11 that has been duplicated is a perfect copy or not.
It appears the African samples and South American samples, while related within continent, are not very closely related to each other.
```{r}
#| fig-column: screen
#| fig-width: 25
#| fig-height: 15
draw(haptype_hrp3_pat1HeatMap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
```
```{r}
pdf("haptype_hrp3_pat1.pdf", useDingbats = F, width = 25, height = 20)
draw(haptype_hrp3_pat1HeatMap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
dev.off()
```
#### Similar samples to 13-11++
Below will get the samples that have a chromosome 11 that is similar to the 13-11++ samples (Deleted hrp3, duplicated sub-telomeric chr11 segment). These new similar samples will be regardless of hrp2/3 deletion status. This will show if the chr11 that has been duplicated is circulating in the general population or is only associated with the samples with HRP3 deletion and chr11 duplication.
```{r}
metaByBioSample_fieldOrIsolate = metaByBioSample %>%
filter(IsFieldSample | "LabIsolate" == site)
jacardDist_gat_filt_forOtherSimilarToPat1 = jacardDist_gat %>%
filter(sample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample |
otherSample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample ) %>%
filter(sample %in% metaByBioSample_fieldOrIsolate$sample,
otherSample %in% metaByBioSample_fieldOrIsolate$sample) %>%
filter(sample %in% samplesCovered$BiologicalSample,
otherSample %in% samplesCovered$BiologicalSample) %>%
# filter(index > 0.99)
filter(index > 0.98)
simSamples = c(unique(c(jacardDist_gat_filt_forOtherSimilarToPat1$sample, jacardDist_gat_filt_forOtherSimilarToPat1$otherSample, allDeletionTypeMeta_hrp3_pat1$BiologicalSample)))
simSamples = simSamples[simSamples != "FCR3"]
jacardDist_gat_filt_simToPat1 = jacardDist_gat %>%
filter(sample %in% simSamples,
otherSample %in% simSamples) %>%
mutate(index = ifelse(is.nan(index), 0, index))
jacardDist_gat_filt_simToPat1_sp = jacardDist_gat_filt_simToPat1 %>%
spread(otherSample, index)
jacardDist_gat_filt_simToPat1_sp_mat = as.matrix(jacardDist_gat_filt_simToPat1_sp[,2:ncol(jacardDist_gat_filt_simToPat1_sp)])
rownames(jacardDist_gat_filt_simToPat1_sp_mat) = jacardDist_gat_filt_simToPat1_sp$sample
```
```{r}
library(circlize)
#['#b2182b','#d6604d','#f4a582','#fddbc7','#f7f7f7','#d1e5f0','#92c5de','#4393c3','#2166ac']
col_fun = colorRamp2(c(min(jacardDist_gat_filt_simToPat1_sp_mat), min(jacardDist_gat_filt_simToPat1_sp_mat) + (1-min(jacardDist_gat_filt_simToPat1_sp_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
jacardDist_gat_filt_simToPat1_sp_mat_noLabs = jacardDist_gat_filt_simToPat1_sp_mat
meta_preferredSample = meta %>%
filter(PreferredSample)
metaSelected_hrp3_pat1 = meta_preferredSample[match(rownames(jacardDist_gat_filt_simToPat1_sp_mat), meta_preferredSample$BiologicalSample), ] %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
select(BiologicalSample, Chr11DupHapCluster))
sample_metadata_withAllDeletionCalls = readr::read_tsv("sample_metadata_withAllDeletionCalls.tsv")
metaSelected_hrp3_pat1 = metaSelected_hrp3_pat1 %>%
left_join(sample_metadata_withAllDeletionCalls %>%
rename(BiologicalSample = sample) %>%
select(BiologicalSample, Pattern)) %>%
mutate(PerfectChr11Copy = case_when(
BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies$s_Sample ~ T,
BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum$s_Sample ~ F,
T ~ NA
))
# %>%
# mutate(hrpCall = ifelse(BiologicalSample %in% previousDeletionCalls$BiologicalSample, hrpCall, "unknown"))
rownames(jacardDist_gat_filt_simToPat1_sp_mat_noLabs) = NULL
colnames(jacardDist_gat_filt_simToPat1_sp_mat_noLabs) = NULL
RowLabs = metaSelected_hrp3_pat1$BiologicalSample
RowLabs[metaSelected_hrp3_pat1$site != "LabIsolate" | is.na(metaSelected_hrp3_pat1$site)] = ""
ColLabs = metaSelected_hrp3_pat1$BiologicalSample
ColLabs[metaSelected_hrp3_pat1$site != "LabIsolate" | is.na(metaSelected_hrp3_pat1$site)] = ""
#RowLabs[metaSelected$country != "Ethiopia"] = ""
rownames(jacardDist_gat_filt_simToPat1_sp_mat_noLabs) = RowLabs
colnames(jacardDist_gat_filt_simToPat1_sp_mat_noLabs) = ColLabs
rowAnnoDf = metaSelected_hrp3_pat1[,c("Pattern", "hrpCall", "PerfectChr11Copy", "country", "region", "secondaryRegion", "Chr11DupHapCluster")] %>% rename(continent = secondaryRegion) %>% as.data.frame()
temp_rowAnnoColors = createColorListFromDf(rowAnnoDf)
temp_rowAnnoColors[["hrpCall"]] = pfhrpsCallColors
temp_rowAnnoColors[["continent"]] = continentColors
temp_rowAnnoColors[["region"]] = rowAnnoColors$region
temp_rowAnnoColors[["Pattern"]] = rowAnnoColorsMod_hrp3DeletionPattern
temp_rowAnnoColors[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
annotationTextSize = 25 ;annotationTitleTextSize = 20;
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = temp_rowAnnoColors,
show_legend = F,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = c("#99999900")
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = temp_rowAnnoColors,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = c("#99999900")
)
haptype_simTo_hrp3_pat1HeatMap = Heatmap(
jacardDist_gat_filt_simToPat1_sp_mat_noLabs,
cluster_columns = T,
col = col_fun,
name = "JacardIndex",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "JacardIndex"
)
)
)
```
Below will get the samples that have a chromosome 11 that is similar to the 13-11++ samples. These new similar samples will be regardless of hrp2/3 deletion status. This will show if the chr11 that has been duplicated is circulating in the general population or is only associated with the samples with HRP3 deletion and chr11 duplication.
It appears that the duplicated chromosome 11 is circulating fairly commonly among South American samples that don't have HRP3 deletion while there doesn't appear to be any of the duplicated chr11 circulating in the African population (though could be a high diversity vs low diversity bias and/or sampling biases given the drastic differences in malaria dynamics in the two continents).
```{r}
#| fig-column: screen
#| fig-width: 30
#| fig-height: 35
draw(haptype_simTo_hrp3_pat1HeatMap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
```
```{r}
pdf("haptype_simTo_hrp3_pat1.pdf", useDingbats = F, width = 30, height = 35)
draw(haptype_simTo_hrp3_pat1HeatMap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
dev.off()
```
## Plotting haplotypes typed per genomic region
Plotting out the variation at the duplicated region, coloring haplotypes by their abundance rank, this visualization will allow interpretation of how similar these haplotypes are here and what the copy looks like within sample (e.g. perfect copy vs variation and how much variation )
### All samples with 13-11++ HRP3 deletion
```{r}
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11, minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample))) %>%
group_by() %>%
filter(sampleCount >= 0.99*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
#filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_dist)
jacardDist_gat_filt_sp_mat_pat1_hc = hclust(dist(jacardDist_gat_filt_sp_mat_pat1))
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_dist_hclust$order])) %>%
# levels = rownames(jacardDist_gat_filt_sp_mat_pat1)[jacardDist_gat_filt_sp_mat_pat1_hc$order])) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name),
expand = c(0,0))+
scale_y_continuous(expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$s_Sample) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
select(BiologicalSample, Chr11DupHapCluster)) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_sp_dist_hclust$order])) %>%
mutate(popid= factor(popid))
```
```{r}
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1 = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
# levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry$p_name),
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry$p_name)))
),
expand = c(0,0)) +
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1 +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapCluster)), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep)+
scale_fill_manual("Chr11DupHapCluster", values = newHaploGroupWithSingletColors, labels = names(newHaploGroupWithSingletColors),
breaks = names(newHaploGroupWithSingletColors)) +
guides(fill = guide_legend(nrow = 4))
```
The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype. Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal (and in this instance would mean the copy on chr11 and chr13 is not a perfect copy).
```{r}
#| fig-column: screen
#| fig-width: 30
#| fig-height: 20
regions_afterHomologous_chr11_filt = regions_afterHomologous_chr11 %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name)))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1 +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -5,
fill = description),
data = regions_afterHomologous_chr11_filt, color = "black") +
scale_fill_manual("Genes\nDescription", values = descriptionColors,
guide = guide_legend(nrow = 5)) +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1)
```
```{r}
pdf("popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_plot.pdf", useDingbats = F, width = 40, height = 30)
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod1)
dev.off()
```
```{r}
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3 = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11, minPopSize = 2)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3 %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample))) %>%
group_by() %>%
filter(sampleCount >= 0.99*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
#filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3 %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_dist_hclust$order])) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$p_name),
expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3 = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$s_Sample) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
select(BiologicalSample, Chr11DupHapCluster)) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3 %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_sp_dist_hclust$order])) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(limits = c(-30, max(c(-9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$p_name))))),
breaks = c(-9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$p_name))),
labels = c("HaploGroup", "continent", levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$p_name)),
expand = c(0,0))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 4)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 4)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapCluster)), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep)+
scale_fill_manual("Chr11DupHapCluster", values = newHaploGroupWithSingletColors, labels = names(newHaploGroupWithSingletColors),
breaks = names(newHaploGroupWithSingletColors)) +
guides(fill = guide_legend(nrow = 4))
regions_afterHomologous_chr11_filt = regions_afterHomologous_chr11 %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3$p_name)))
yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$s_Sample)
yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3[yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 %!in% c("HB3", "Santa-Lucia-Salvador-I", "SD01")] = ""
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 +
scale_y_continuous(labels = yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3,
breaks = 1:length(yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3),
expand = c(0,0)) +
theme(axis.text.x = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank(),
panel.border = element_blank(),
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3_priorToGeneInfo = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3 +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -7,
fill = description),
data = regions_afterHomologous_chr11_filt, color = "black") +
geom_text(
aes(y = as.numeric(BiologicalSample),
x = -10,
label = BiologicalSample),
hjust = 1,
#data = tibble(BiologicalSample = factor(c("HB3", "Santa-Lucia-Salvador-I", "SD01"), levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$s_Sample)))
data = tibble(BiologicalSample = factor(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$s_Sample, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod3_withCountry$s_Sample)))
) +
scale_fill_manual("Genes\nDescription", values = descriptionColors,
guide = guide_legend(nrow = 4))
```
The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype. Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal (and in this instance would mean the copy on chr11 and chr13 is not a perfect copy).
```{r}
#| fig-column: screen
#| fig-width: 30
#| fig-height: 20
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3)
```
##### Collapsing parasites by same haplotypes
```{r}
pdf("popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_plot_mod3_onlyVariableSites.pdf", useDingbats = F, width = 25, height = 20)
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3)
dev.off()
pdf("popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_plot_mod3_onlyVariableSites_noGeneInfo.pdf", useDingbats = F, width = 15, height = 12.5)
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod3_priorToGeneInfo)
dev.off()
```
```{r}
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
group_by(s_Sample) %>%
summarise(p_name_count = length(unique(p_name)),
p_name_meanCOI = mean(uniqHaps)) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
rename(s_Sample = BiologicalSample))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness %>%
#filter(s_Sample %!in% c("HB3", "QV0040-C", "IGS-CBD-010")) %>%
#filter(hcclustSize > 2, newClusterName != 9) %>%
#filter(hcclustSize > 1, newClusterName != 9) %>%
filter(hcclustSize > 1) %>%
arrange(desc(p_name_count), p_name_meanCOI) %>%
group_by(newClusterName) %>%
mutate(groupID = row_number()) %>%
filter(groupID == 1) %>%
left_join(meta_preferredSample %>%
select(BiologicalSample, secondaryRegion) %>%
rename(s_Sample = BiologicalSample))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt %>%
mutate(secondaryRegion = factor(secondaryRegion, levels = c("S_AMERICA", "AFRICA", "ASIA"))) %>%
arrange(secondaryRegion, desc(hcclustSize)) %>%
mutate(s_Sample = factor(s_Sample, levels = .$s_Sample))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4 = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
filter(s_Sample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt$s_Sample), minPopSize = 2)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4 %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample))) %>%
group_by() %>%
filter(sampleCount >= 0.99*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist)
jacardDist_gat_filt_sp_mat_pat1_hc = hclust(dist(jacardDist_gat_filt_sp_mat_pat1))
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4 %>%
mutate(s_Sample = factor(s_Sample,
levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt$s_Sample))) %>%
# levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust$order])) %>%
# levels = rownames(jacardDist_gat_filt_sp_mat_pat1)[jacardDist_gat_filt_sp_mat_pat1_hc$order])) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$p_name),
expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4 = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$s_Sample) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4 %>%
mutate(s_Sample = factor(s_Sample,
levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt$s_Sample))) %>%
# mutate(s_Sample = factor(s_Sample,
# levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust$order])) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(limits = c(-30, max(c(-9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry$p_name))))),
breaks = c(-9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry$p_name))),
labels = c("HaploGroup", "continent", levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry$p_name)),
expand = c(0,0))
k_groups = nrow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_regionCompletionness_filt);
h_groups = 1.1;
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups = cutree(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust, k = k_groups)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend <- as.dendrogram(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend <- color_labels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend, k = k_groups)
plot(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups = cutree(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust, h = h_groups)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend <- as.dendrogram(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend <- color_labels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend, h = h_groups)
plot(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_dend)
jacardDist_gat_filt_sp_mat_pat1_hc_groups = cutree(jacardDist_gat_filt_sp_mat_pat1_hc, k = k_groups)
jacardDist_gat_filt_sp_mat_pat1_hc_dend <- as.dendrogram(jacardDist_gat_filt_sp_mat_pat1_hc)
jacardDist_gat_filt_sp_mat_pat1_hc_dend <- color_labels(jacardDist_gat_filt_sp_mat_pat1_hc_dend, k = k_groups)
plot(jacardDist_gat_filt_sp_mat_pat1_hc_dend)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df = tibble(
BiologicalSample = names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups),
hcclust = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups
) %>%
mutate(BiologicalSample =factor(BiologicalSample, levels = levels(meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$BiologicalSample))) %>%
group_by(hcclust) %>%
mutate(hcclustSize = n())
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_biggerGroups = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df %>% filter(hcclustSize != 1) %>% select(hcclust) %>% unique()
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_singletonGroups = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df %>% filter(hcclustSize == 1) %>% select(hcclust) %>% unique()
nonSingletonGroupsColors = scheme$hex(nrow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df %>% select(hcclust) %>% unique()))
names(nonSingletonGroupsColors) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_biggerGroups$hcclust
nonSingletonGroupsColors_singleton = scheme$hex(nrow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df %>% select(hcclust) %>% unique()))
names(nonSingletonGroupsColors_singleton) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_singletonGroups$hcclust
haploGroupColors = c(nonSingletonGroupsColors, nonSingletonGroupsColors_singleton)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_groupCounts = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df %>%
select(hcclust, hcclustSize) %>%
ungroup() %>%
unique() %>%
arrange(desc(hcclustSize)) %>%
mutate(hcclust = as.character(hcclust),newClusterName = row_number()) %>%
left_join(tibble(
hcclust = names(haploGroupColors),
colors = unname(haploGroupColors)
))
newHaploGroupColors = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_groupCounts$colors
names(newHaploGroupColors)= popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_groupCounts$newClusterName
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df %>%
left_join(
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df_groupCounts %>%
mutate(hcclust = as.integer(hcclust))
) %>%
left_join(
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>% ungroup() %>% select(BiologicalSample, hcclustSize) %>% rename(originalGroupSize = hcclustSize)
) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$BiologicalSample)))
# jacardDist_gat_filt_sp_mat_pat1_hc_groups_df = tibble(
# BiologicalSample = names(jacardDist_gat_filt_sp_mat_pat1_hc_groups),
# hcclust = jacardDist_gat_filt_sp_mat_pat1_hc_groups
# ) %>%
# mutate(BiologicalSample =factor(BiologicalSample, levels = levels(meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$BiologicalSample)))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))]) ) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.3,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 4)) +
# ggnewscale::new_scale_fill() +
# geom_rect(aes(xmin= -5, xmax = -9.5,
# ymin = as.numeric(BiologicalSample) - 0.5,
# ymax = as.numeric(BiologicalSample) + 0.3,
# fill = factor(newClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df)+
# # fill = factor(hcclust)), color = "black", data = jacardDist_gat_filt_sp_mat_pat1_hc_groups_df)+
# # scale_fill_manual("HaploGroup", values = scheme$hex(length(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups)))) +
# #scale_fill_manual("Chr11DupHapCluster", values = haploGroupColors, labels = names(haploGroupColors), breaks = names(haploGroupColors)) +
# scale_fill_manual("Chr11DupHapCluster", values = newHaploGroupColors, labels = names(newHaploGroupColors),
# breaks = names(newHaploGroupColors)) +
geom_text(aes(
x = -9.5,
y = as.numeric(BiologicalSample) - 0.5 + 0.4,
label = paste0("n=", originalGroupSize)
), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_sp_dist_hclust_groups_df)+
guides(fill = guide_legend(nrow = 4))
regions_afterHomologous_chr11_filt = regions_afterHomologous_chr11 %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4$p_name)))
yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry$s_Sample)
# yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4[yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 %!in% c("HB3", "Santa-Lucia-Salvador-I", "SD01")] = ""
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 +
scale_y_continuous(labels = yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4,
breaks = 1:length(yLabels_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4),
expand = c(0,0)) +
theme(axis.text.x = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank(),
panel.border = element_blank(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black", linewidth = 1)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4_priorToGeneInfo = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4 +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -1,
fill = description),
data = regions_afterHomologous_chr11_filt, color = "black") +
geom_text(
aes(y = as.numeric(BiologicalSample),
x = -10,
label = BiologicalSample),
hjust = 1,
data = tibble(BiologicalSample = factor(c("HB3", "Santa-Lucia-Salvador-I", "SD01"), levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_mod4_withCountry$s_Sample)))
) +
scale_fill_manual("Genes\nDescription", values = descriptionColors,
guide = guide_legend(nrow = 4))
```
The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype. Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal (and in this instance would mean the copy on chr11 and chr13 is not a perfect copy).
```{r}
#| fig-column: screen
#| fig-width: 30
#| fig-height: 20
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4)
```
```{r}
pdf("popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_plot_mod4_onlyVariableSites.pdf", useDingbats = F, width = 25, height = 15)
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4)
dev.off()
pdf("popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_plot_mod4_onlyVariableSites_noGeneInfo.pdf", useDingbats = F, width = 15, height = 6)
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_withCountry_plot_mod4_priorToGeneInfo)
dev.off()
```
#### Perfect copies
```{r}
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
filter(s_Sample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies$s_Sample) , minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample))) %>%
group_by() %>%
filter(sampleCount > 0.9*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_dist_hclust$order]))%>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$p_name),
expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$s_Sample) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_sp_dist_hclust$order])) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry$p_name)))
# levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry$p_name)
),
expand = c(0,0)) +
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies =
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$s_Sample) %>%
mutate(BiologicalSample = factor(as.character(BiologicalSample), levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies$s_Sample))) %>%
mutate(Chr11DupHapClusterName = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, width = 2, pad = "0")))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColorsDf = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies %>%
select(Chr11DupHapClusterName, colors) %>%
unique() %>%
arrange(Chr11DupHapClusterName)
#
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColors = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColorsDf$colors
names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColors) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColorsDf$Chr11DupHapClusterName
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies)+
# fill = factor(hcclust)), color = "black", data = jacardDist_gat_filt_sp_mat_pat1_hc_groups_df)+
# scale_fill_manual("HaploGroup", values = scheme$hex(length(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups)))) +
#scale_fill_manual("Chr11DupHapCluster", values = haploGroupColors, labels = names(haploGroupColors), breaks = names(haploGroupColors)) +
scale_fill_manual("Chr11DupHapCluster", values = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColors, labels = names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColors),
breaks = names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_Chr11DupHapClusterColors)) +
guides(fill = guide_legend(nrow = 4))
```
The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal (and in this instance would mean the copy on chr11 and chr13 is not a perfect copy)
```{r}
#| fig-column: screen
#| fig-width: 40
#| fig-height: 35
regions_afterHomologous_chr11_filt = regions_afterHomologous_chr11 %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name)))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -5,
fill = description),
data = regions_afterHomologous_chr11_filt, color = "black") +
scale_fill_manual("Genes\nDescription", values = descriptionColors,
guide = guide_legend(nrow = 5)) +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot)
```
```{r}
pdf("popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_plot.pdf", useDingbats = F, width = 40, height = 35)
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_closeToPerfectCopies_withCountry_plot + labs(title = "Perfect Copies"))
dev.off()
```
#### Divergent copies
```{r}
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
filter(s_Sample %!in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies$s_Sample) , minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies %>%
group_by() %>%
filter(samp_n > 0.9*max(samp_n)) %>%
group_by(s_Sample, p_name) %>%
#filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_dist)
nameOrderFromforClustering = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust$order]
orderForDivergentCopy = nameOrderFromforClustering[nameOrderFromforClustering %in% rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_mat)]
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies %>%
mutate(s_Sample = factor(s_Sample,
# levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_dist_hclust$order]))%>%
levels = orderForDivergentCopy)) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$p_name),
expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$s_Sample) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies %>%
mutate(s_Sample = factor(s_Sample,
# levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_sp_dist_hclust$order])) %>%
levels = orderForDivergentCopy)) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies =
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$s_Sample) %>%
mutate(BiologicalSample = factor(as.character(BiologicalSample), levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies$s_Sample))) %>%
mutate(Chr11DupHapClusterName = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, width = 2, pad = "0"))) %>%
arrange(BiologicalSample)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColorsDf = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies %>%
select(Chr11DupHapClusterName, colors) %>%
unique() %>%
arrange(Chr11DupHapClusterName)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColors = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColorsDf$colors
names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColors) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColorsDf$Chr11DupHapClusterName
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry$p_name)))
# levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry$p_name)
),
expand = c(0,0))+
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot+
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies)+
# fill = factor(hcclust)), color = "black", data = jacardDist_gat_filt_sp_mat_pat1_hc_groups_df)+
# scale_fill_manual("HaploGroup", values = scheme$hex(length(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups)))) +
#scale_fill_manual("Chr11DupHapCluster", values = haploGroupColors, labels = names(haploGroupColors), breaks = names(haploGroupColors)) +
scale_fill_manual("Chr11DupHapCluster", values = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColors, labels = names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColors),
breaks = names(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_Chr11DupHapClusterColors)) +
guides(fill = guide_legend(nrow = 4))
```
The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal (and in this instance would mean the copy on chr11 and chr13 is not a perfect copy)
```{r}
#| fig-column: screen
#| fig-width: 30
#| fig-height: 20
regions_afterHomologous_chr11_filt = regions_afterHomologous_chr11 %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep$p_name)))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -1,
fill = description),
data = regions_afterHomologous_chr11_filt, color = "black") +
scale_fill_manual("Genes\nDescription", values = descriptionColors,
guide = guide_legend(nrow = 5)) +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot)
```
```{r}
pdf("popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_plot.pdf", useDingbats = F, width = 40, height = 30)
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_divergentCopies_withCountry_plot + labs(title = "Divergent Copies"))
dev.off()
```
### Sub set
### SD01, HB3, Santa-Lucia-Salvador-I
```{r}
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11 %>%
filter(s_Sample %in% c("HB3", "SD01", "Santa-Lucia-Salvador-I")) , minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample)))%>%
group_by() %>%
filter(sampleCount > 0.9*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_mat) = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_mat)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_mat)[popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_sp_dist_hclust$order]))
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$p_name)),
# labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$p_name),
labels = rep("", length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$p_name))),
expand = c(0,0))
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$popid))
previousColors["-1"] = "grey0";
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates%>%
mutate(popid= factor(popid)), colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$p_name)),
# labels = levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$p_name),
labels = rep("", length(levels(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates$p_name))),
expand = c(0,0))
```
The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal (and in this instance would mean the copy on chr11 and chr13 is not a perfect copy)
It appears that SD01 and Santa-Lucia-Salvador-I have perfect copies of chr11 on chr11 and chr13 while HB3 has a divergent copy (which is confirmed with the nanopore assembly)
Interestingly enough, the Santa-Lucia-Salvador-I chr11 duplicated region appears to be one of the chr11 in HB3.
```{r}
#| fig-column: screen
#| fig-width: 30
#| fig-height: 5
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 1)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -1,
fill = description),
data = regions_afterHomologous_chr11_filt, color = "black") +
scale_fill_manual("Genes\nDescription", values = descriptionColors,
guide = guide_legend(nrow = 5)) +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30),
axis.text.y = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot)
```
```{r}
pdf("popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot.pdf", useDingbats = F, width = 40, height = 7.5)
print(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_LabIsolates_plot)
dev.off()
```
# Shared Region between chr11 and chr13
The data on the 15.2kb duplicated region between chromosome 11 and 13.
```{r}
excludeRegions = c("Pf3D7_11_v3-1919633-1920323-for__var-6",
"Pf3D7_11_v3-1920483-1921173-for__var-3",
"Pf3D7_11_v3-1920483-1921173-for__var-4",
"Pf3D7_11_v3-1920483-1921173-for__var-5",
"Pf3D7_11_v3-1920483-1921173-for__var-6",
"Pf3D7_11_v3-1920483-1921173-for__var-7",
"Pf3D7_11_v3-1928369-1928869-for__var-3",
"Pf3D7_11_v3-1928619-1929119-for__var-3")
regions_homologousRegion = regions %>%
filter("shared" == homologousRegion) %>%
filter(`#chrom` == "Pf3D7_11_v3") %>%
filter(name %!in% excludeRegions)
popClustering_filt_hrp3_pat1_regions_homologousRegion = popClustering_filt_hrp3_pat1 %>%
filter(p_name %in% regions_homologousRegion$genomicID)
popClustering_filt_hrp3_pat1_regions_homologousRegion = popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
group_by(s_Sample, p_name) %>%
mutate(uniqHaps= n())
popClustering_filt_hrp3_pat1_regions_homologousRegion_uniqSum = popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
group_by(s_Sample) %>%
mutate(targets = length(unique(genomicID))) %>%
group_by(s_Sample, targets, uniqHaps) %>%
count() %>%
mutate(freq = n/targets)
popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum = popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
mutate(marker = uniqHaps == 1) %>%
group_by(s_Sample) %>%
summarise(conserved = sum(marker),
targets = length(unique(genomicID))) %>%
mutate(conservedID = conserved/targets)
conservedCutOff = 0.99
popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_closeToPerfectCopies = popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum %>%
filter(conservedID > conservedCutOff)
popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_cutOff = popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
group_by() %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)
popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_cutOffByRegion = popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
left_join(metaByBioSample %>%
rename(s_Sample = sample)) %>%
group_by(region) %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)
popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_cutOffByContinent = popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum %>%
mutate(marker = conservedID > conservedCutOff) %>%
left_join(metaByBioSample %>%
rename(s_Sample = sample)) %>%
group_by(secondaryRegion) %>%
summarise(perfectDuplication = sum(marker),
totalSamps = length(unique(s_Sample))) %>%
mutate(perfectCopyFreq = perfectDuplication/totalSamps)
```
The number of samples with perfect copies
```{r}
create_dt(popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_cutOff)
```
The number of samples with perfect copies broken down by regions
```{r}
create_dt(popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_cutOffByRegion)
```
The number of samples with perfect copies broken down by continent.
```{r}
create_dt(popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_cutOffByContinent)
popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_meanId = popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum %>%
filter(conservedID <= conservedCutOff) %>%
summarise(meanID = mean(conservedID),
minID = min(conservedID),
sdID = sd(conservedID))
```
The breakdown of level of divergence in the samples with divergent samples.
```{r}
create_dt(popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_meanId)
```
## Population analysis of chr11/chr13 shared region
Calculating the population of the haplotypes of the shared region on chr 11/chr13
```{r}
popClustering_filt_regions_homologousRegion = popClustering %>%
filter(genomicID %!in% erroneousRegions) %>%
filter(p_name %in% regions_homologousRegion$genomicID)
popClustering_filt_regions_homologousRegion_tarCounts = popClustering_filt_regions_homologousRegion %>%
group_by(s_Sample) %>%
summarise(tarCounts = length(unique(p_name)))
popClustering_filt_regions_homologousRegion_tarCounts_filt = popClustering_filt_regions_homologousRegion_tarCounts %>%
filter(tarCounts >= 0.80 * max(tarCounts) |
s_Sample %in% previousDeletionCalls$BiologicalSample)
popClustering_filt_regions_homologousRegion_sampCounts = popClustering_filt_regions_homologousRegion %>%
group_by(p_name) %>%
summarise(sampCounts = length(unique(s_Sample)))
write_tsv(popClustering_filt_regions_homologousRegion %>%
filter(s_Sample %in% popClustering_filt_regions_homologousRegion_tarCounts_filt$s_Sample) %>%
group_by() %>%
select(s_Sample, p_name, h_popUID, c_AveragedFrac),
"popClustering_filt_regions_homologousRegion.tab.txt.gz")
```
```{bash, eval = F}
elucidator doPairwiseComparisonOnHapsSharingDev --tableFnp popClustering_filt_regions_homologousRegion.tab.txt.gz --sampleCol s_Sample --targetNameCol p_name --popIDCol h_popUID --relAbundCol c_AveragedFrac --numThreads 14 --dout pairwiseComps_regions_homologousRegion --verbose --overWriteDir --metaFnp metaByBioSample_outwithHrpCalls.tab.txt --metaFieldsToCalcPopDiffs country,region,secondaryRegion,HRP3_deletionPattern --writeOutDistMatrices
```
```{r}
#jacardDist = readr::read_tsv("pairwiseComps_regions_homologousRegion/percOfTarSharingAtLeastOneHap.tab.txt.gz", col_names = F)
jacardDist_homologousRegion = readr::read_tsv("pairwiseComps_regions_homologousRegion/jacardByHapsTarShared.tab.txt.gz", col_names = F)
jacardDist_homologousRegionSamps = readr::read_tsv("pairwiseComps_regions_homologousRegion/sampleNames.tab.txt", col_names = "samples")
colnames(jacardDist_homologousRegion) = jacardDist_homologousRegionSamps$samples
jacardDist_homologousRegion$sample = jacardDist_homologousRegionSamps$samples
# jacardDist_homologousRegion_filt = jacardDist_homologousRegion %>%
# filter(sample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample)
jacardDist_homologousRegion_filt = jacardDist_homologousRegion[jacardDist_homologousRegion$sample %in% allDeletionTypeMeta_hrp3_pat1$BiologicalSample,c(allDeletionTypeMeta_hrp3_pat1$BiologicalSample, "sample")]
jacardDist_homologousRegion_gat = jacardDist_homologousRegion_filt %>%
gather(otherSample, index,1:(ncol(.) - 1))
jacardDist_homologousRegion_gat_filt = jacardDist_homologousRegion_gat %>%
filter(sample %fin% allDeletionTypeMeta_hrp3_pat1$BiologicalSample,
otherSample %fin% allDeletionTypeMeta_hrp3_pat1$BiologicalSample)
jacardDist_homologousRegion_gat_filt_sp = jacardDist_homologousRegion_gat_filt %>%
spread(otherSample, index)
jacardDist_homologousRegion_gat_filt_sp_mat = as.matrix(jacardDist_homologousRegion_gat_filt_sp[,2:ncol(jacardDist_homologousRegion_gat_filt_sp)])
rownames(jacardDist_homologousRegion_gat_filt_sp_mat) = jacardDist_homologousRegion_gat_filt_sp$sample
```
```{r}
library(circlize)
#col_fun = colorRamp2(c(0, 0.5, 1), c(heat.colors(3)))
col_fun = colorRamp2(c(min(jacardDist_homologousRegion_gat_filt_sp_mat), min(jacardDist_homologousRegion_gat_filt_sp_mat) + (1-min(jacardDist_homologousRegion_gat_filt_sp_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
jacardDist_homologousRegion_gat_filt_sp_mat_noLabs = jacardDist_homologousRegion_gat_filt_sp_mat
meta_preferredSample = meta %>%
filter(PreferredSample)
metaSelected_hrp3_pat1 = meta_preferredSample[match(rownames(jacardDist_homologousRegion_gat_filt_sp_mat), meta_preferredSample$BiologicalSample), ]
metaSelected_hrp3_pat1 = metaSelected_hrp3_pat1 %>%
mutate(PerfectChr11Copy = BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies$s_Sample) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
mutate(newClusterName = ifelse(hcclustSize == 1, "singlet", as.character(stringr::str_pad(newClusterName, width = 2, pad = "0")))) %>%
mutate(BiologicalSample = as.character(BiologicalSample)) %>%
select(BiologicalSample, newClusterName))
rownames(jacardDist_homologousRegion_gat_filt_sp_mat_noLabs) = NULL
colnames(jacardDist_homologousRegion_gat_filt_sp_mat_noLabs) = NULL
RowLabs = metaSelected_hrp3_pat1$BiologicalSample
RowLabs[metaSelected_hrp3_pat1$site != "LabIsolate" | is.na(metaSelected_hrp3_pat1$site)] = ""
ColLabs = metaSelected_hrp3_pat1$BiologicalSample
ColLabs[metaSelected_hrp3_pat1$site != "LabIsolate" | is.na(metaSelected_hrp3_pat1$site)] = ""
#RowLabs[metaSelected$country != "Ethiopia"] = ""
rownames(jacardDist_homologousRegion_gat_filt_sp_mat_noLabs) = RowLabs
colnames(jacardDist_homologousRegion_gat_filt_sp_mat_noLabs) = ColLabs
rowAnnoDf = metaSelected_hrp3_pat1[,c("hrpCall", "PerfectChr11Copy", "country", "region", "secondaryRegion", "newClusterName")] %>% rename(continent = secondaryRegion,
Chr11DupHapCluster = newClusterName) %>% as.data.frame()
annotationTextSize = 25 ;annotationTitleTextSize = 20;
rowAnnoColors_mod = rowAnnoColors
rowAnnoColors_mod[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
show_legend = F,
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
gp = gpar(col = "grey10")
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
gp = gpar(col = "grey10")
)
haptype_hrp3_regions_homologousRegion_pat1HeatMap = Heatmap(
jacardDist_homologousRegion_gat_filt_sp_mat_noLabs,
cluster_columns = T,
col = col_fun,
name = "JacardIndex",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "JacardIndex"
)
)
)
```
```{r}
#| fig-column: screen
#| fig-width: 25
#| fig-height: 15
draw(haptype_hrp3_regions_homologousRegion_pat1HeatMap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
```
```{r}
pdf("haptype_hrp3_regions_homologousRegion_pat1HeatMap.pdf", useDingbats = F, width = 25, height = 20)
draw(haptype_hrp3_regions_homologousRegion_pat1HeatMap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
dev.off()
```
### Plotting haplotypes
Plotting out the variation at the duplicated region, coloring haplotypes by their abundance rank, this visualization will allow interpretation of how similar these haplotypes are here and what the copy looks like within sample (e.g. perfect copy vs variation and how much variation )
### All
```{r}
regions_homologousRegion = regions_homologousRegion %>%
mutate(description = case_when(
grepl("extraField0=NA", extraField0) ~ "intergenic",
T ~ gsub("\\]", "", gsub(".*description=", "", extraField0))
) )
descriptionColors_homologousRegion = scheme$hex(length(regions_homologousRegion$description %>% unique()))
names(descriptionColors_homologousRegion) = regions_homologousRegion$description %>% unique()
descriptionColors_homologousRegion["intergenic"] = c("#FF000000")
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_homologousRegion, minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample)))%>%
group_by() %>%
filter(sampleCount > 0.9*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_mat) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_mat)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_dist_hclust$order])) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$p_name),
expand = c(0,0)) +
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$s_Sample)
)
meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$s_Sample) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_dist_hclust$order])) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$p_name)))
# levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$p_name)
),
expand = c(0,0))+
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry =
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$s_Sample) %>%
mutate(BiologicalSample = factor(as.character(BiologicalSample), levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$s_Sample))) %>%
mutate(Chr11DupHapClusterName = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, width = 2, pad = "0"))) %>%
arrange(BiologicalSample)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColorsDf = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry %>%
select(Chr11DupHapClusterName, colors) %>%
unique() %>%
arrange(Chr11DupHapClusterName)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColors = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColorsDf$colors
names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColors) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColorsDf$Chr11DupHapClusterName
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry)+
# fill = factor(hcclust)), color = "black", data = jacardDist_gat_filt_sp_mat_pat1_hc_groups_df)+
# scale_fill_manual("HaploGroup", values = scheme$hex(length(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups)))) +
#scale_fill_manual("Chr11DupHapCluster", values = haploGroupColors, labels = names(haploGroupColors), breaks = names(haploGroupColors)) +
scale_fill_manual("Chr11DupHapCluster", values = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColors, labels = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColors),
breaks = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_Chr11DupHapClusterColors)) +
guides(fill = guide_legend(nrow = 4))
```
The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal
```{r}
#| fig-column: screen
#| fig-width: 30
#| fig-height: 20
regions_homologousRegion_filt = regions_homologousRegion %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry$p_name)))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot_final = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -10,
fill = description),
data = regions_homologousRegion_filt, color = "black") +
scale_fill_manual("Genes\nDescription", values = descriptionColors_homologousRegion,
guide = guide_legend(nrow = 2) ) +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot_final )
```
```{r}
pdf("popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_plot.pdf",useDingbats = F,width = 30,height = 25)
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_withCountry_plot_final)
dev.off()
```
#### Perfect copies
```{r}
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
filter(s_Sample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_closeToPerfectCopies$s_Sample) , minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample)))%>%
group_by() %>%
filter(sampleCount > 0.9*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_mat) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_mat)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_dist_hclust$order]))%>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies$p_name),
expand = c(0,0))+
scale_y_continuous(expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies$s_Sample) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_sp_dist_hclust$order])) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$p_name)))
# levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$p_name)
),
expand = c(0,0))+
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry =
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$s_Sample) %>%
mutate(BiologicalSample = factor(as.character(BiologicalSample), levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$s_Sample))) %>%
mutate(Chr11DupHapClusterName = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, width = 2, pad = "0"))) %>%
arrange(BiologicalSample)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColorsDf = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry %>%
select(Chr11DupHapClusterName, colors) %>%
unique() %>%
arrange(Chr11DupHapClusterName)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColors = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColorsDf$colors
names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColors) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColorsDf$Chr11DupHapClusterName
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot+
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry)+
# fill = factor(hcclust)), color = "black", data = jacardDist_gat_filt_sp_mat_pat1_hc_groups_df)+
# scale_fill_manual("HaploGroup", values = scheme$hex(length(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups)))) +
#scale_fill_manual("Chr11DupHapCluster", values = haploGroupColors, labels = names(haploGroupColors), breaks = names(haploGroupColors)) +
scale_fill_manual("Chr11DupHapCluster", values = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColors, labels = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColors),
breaks = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_Chr11DupHapClusterColors)) +
guides(fill = guide_legend(nrow = 4))
```
The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal
```{r}
#| fig-column: screen
#| fig-width: 30
#| fig-height: 20
regions_homologousRegion_filt = regions_homologousRegion %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry$p_name)))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot_final = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -1,
fill = description),
data = regions_homologousRegion_filt, color = "black") +
scale_fill_manual("Genes\nDescription", values = descriptionColors_homologousRegion,
guide = guide_legend(nrow = 2))+
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot_final)
```
```{r}
pdf("popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_plot.pdf", useDingbats = F, width = 30, height = 25)
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_closeToPerfectCopies_withCountry_plot_final)
dev.off()
```
#### Divergent copies
Divergent copies of the shared region
```{r}
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
filter(s_Sample %!in% popClustering_filt_hrp3_pat1_regions_homologousRegion_conservedSum_closeToPerfectCopies$s_Sample) , minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies %>%
group_by() %>%
filter(samp_n > 0.9*max(samp_n)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_mat) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_mat)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_dist)
nameOrderFromAll = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_sp_dist_hclust$order]
orderForDivergentCopy = nameOrderFromAll[nameOrderFromAll %in% rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_mat)]
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies %>%
mutate(s_Sample = factor(s_Sample,
# levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_dist_hclust$order]))%>%
levels = orderForDivergentCopy)) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies$p_name),
expand = c(0,0))+
scale_y_continuous(expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies$s_Sample) %>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies %>%
mutate(s_Sample = factor(s_Sample,
# levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_sp_dist_hclust$order])) %>%
levels = orderForDivergentCopy)) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$p_name)))
# levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$p_name)
),
expand = c(0,0))+
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry =
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$s_Sample) %>%
mutate(BiologicalSample = factor(as.character(BiologicalSample), levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$s_Sample))) %>%
mutate(Chr11DupHapClusterName = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, width = 2, pad = "0"))) %>%
arrange(BiologicalSample)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColorsDf = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry %>%
select(Chr11DupHapClusterName, colors) %>%
unique() %>%
arrange(Chr11DupHapClusterName)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColors = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColorsDf$colors
names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColors) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColorsDf$Chr11DupHapClusterName
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry)+
# fill = factor(hcclust)), color = "black", data = jacardDist_gat_filt_sp_mat_pat1_hc_groups_df)+
# scale_fill_manual("HaploGroup", values = scheme$hex(length(unique(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups)))) +
#scale_fill_manual("Chr11DupHapCluster", values = haploGroupColors, labels = names(haploGroupColors), breaks = names(haploGroupColors)) +
scale_fill_manual("Chr11DupHapCluster", values = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColors, labels = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColors),
breaks = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_Chr11DupHapClusterColors)) +
guides(fill = guide_legend(nrow = 4))
```
The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal
```{r}
#| fig-column: screen
#| fig-width: 30
#| fig-height: 20
regions_homologousRegion_filt = regions_homologousRegion %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry$p_name)))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot_final = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -10,
fill = description),
data = regions_homologousRegion_filt, color = "black") +
scale_fill_manual(values = descriptionColors_homologousRegion,
guide = guide_legend(nrow = 2)) +
labs(fill = "Genes\nDescription") +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot_final)
```
```{r}
pdf("popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_plot.pdf", useDingbats = F, width = 30, height = 30)
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_divergentCopies_withCountry_plot_final)
dev.off()
```
#### Perfect chr11 copies
The shared region of the strains with perfect chr11 copies.
```{r}
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies = HaplotypeRainbows::prepForRainbow(
popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
filter(
s_Sample %in% popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_conservedSum_closeToPerfectCopies$s_Sample
),
minPopSize = 1
)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies %>%
group_by() %>%
# filter(samp_n > 0.9*max(samp_n)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_mat) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_mat)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist_hclust_orderDf =
tibble(
BiologicalSample = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist_hclust$order]
) %>%
mutate(byGenomicRegionHclustOrder = row_number())
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
arrange(newClusterName)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_select = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies$s_Sample) %>%
left_join(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist_hclust_orderDf) %>%
left_join(meta_preferredSample %>%
select(BiologicalSample, country, subRegion, region, secondaryRegion)) %>%
arrange(Chr11DupHapCluster, subRegion, country, byGenomicRegionHclustOrder)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies %>%
mutate(s_Sample = factor(s_Sample,
#levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist_hclust$order]))%>%
levels = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_select$BiologicalSample) ) %>%
mutate(popid = ifelse(maxPopid == 1, -1, popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies$p_name)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies$p_name),
expand = c(0,0))+
scale_y_continuous(expand = c(0,0))
meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies = meta_preferredSample %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies$s_Sample) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_select %>%
select(BiologicalSample, newClusterName))%>%
mutate(BiologicalSample = factor(BiologicalSample, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies$s_Sample)))
allColors = c(); for(name in names(rowAnnoColors)){ allColors = c(allColors, rowAnnoColors[[name]])}
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies$popid))
previousColors["-1"] = "grey0";
allColors = c(allColors, previousColors)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies %>%
# mutate(s_Sample = factor(s_Sample,
# levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_sp_dist_hclust$order])) %>%
mutate(s_Sample = factor(s_Sample,
levels = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_select$BiologicalSample)) %>%
mutate(popid= factor(popid))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = c(-19.5 + 2.25, -14.5 + 2.25, -9.5 + 2.25, -4.5 + 2.25, 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$p_name))),
labels = c("Chr11DupHapCluster", "continent", "region", "country",
rep("", length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$p_name)))
# levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$p_name)
),
expand = c(0,0))+
scale_y_continuous(
expand = c(0, 0),
breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$s_Sample)
)
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry =
popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
filter(BiologicalSample %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$s_Sample) %>%
mutate(BiologicalSample = factor(as.character(BiologicalSample), levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$s_Sample))) %>%
mutate(Chr11DupHapClusterName = ifelse(hcclustSize == 1, "singlet", stringr::str_pad(newClusterName, width = 2, pad = "0"))) %>%
arrange(BiologicalSample)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColorsDf = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry %>%
select(Chr11DupHapClusterName, colors) %>%
unique() %>%
arrange(Chr11DupHapClusterName)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColors = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColorsDf$colors
names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColors) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColorsDf$Chr11DupHapClusterName
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= 0, xmax = -4.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = country), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies) +
scale_fill_manual("country", values = rowAnnoColors[["country"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -5, xmax = -9.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = region), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies) +
scale_fill_manual("region", values = rowAnnoColors[["region"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -10, xmax = -14.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = secondaryRegion), color = "black", data = meta_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies)+
scale_fill_manual("Continent", values = rowAnnoColors[["continent"]]) +
guides(fill = guide_legend(nrow = 3)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin= -15, xmax = -19.5,
ymin = as.numeric(BiologicalSample) - 0.5,
ymax = as.numeric(BiologicalSample) + 0.5,
fill = factor(Chr11DupHapClusterName)), color = "black", data = popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df_in_popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry)+
scale_fill_manual("Chr11DupHapCluster", values = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColors, labels = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColors),
breaks = names(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_Chr11DupHapClusterColors)) +
guides(fill = guide_legend(nrow = 4))
```
The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal
```{r}
#| fig-column: screen
#| fig-width: 30
#| fig-height: 20
regions_homologousRegion_filt = regions_homologousRegion %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry$p_name)))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot_final = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -10,
fill = description),
data = regions_homologousRegion_filt, color = "black") +
scale_fill_manual(values = descriptionColors_homologousRegion,
guide = guide_legend(nrow = 2)) +
labs(fill = "Genes\nDescription") +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot_final)
```
```{r}
pdf("popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_plot.pdf", useDingbats = F, width = 30, height = 30)
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_perfectChr11Copies_withCountry_plot_final)
dev.off()
```
### Sub set
### SD01, HB3, Santa-Lucia-Salvador-I
```{r}
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates = HaplotypeRainbows::prepForRainbow(popClustering_filt_hrp3_pat1_regions_homologousRegion %>%
filter(s_Sample %in% c("HB3", "SD01", "Santa-Lucia-Salvador-I")) , minPopSize = 1)
# select just the major haplotypes and cluster based on the sharing between
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample)))%>%
group_by()%>%
filter(sampleCount > 0.9*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_mat = as.matrix(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp[,2:ncol(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp)])
rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_mat) = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp$s_Sample
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_dist = dist(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_mat)
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_dist_hclust = hclust(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_mat)[popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sp_dist_hclust$order]))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name)),
#labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name),
labels = rep("", length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name))),
expand = c(0,0))+
scale_y_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$s_Sample),
expand = c(0,0))
previousColors = unique(ggplot_build(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot)$data[[1]][["fill"]])
names(previousColors) = sort(unique(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$popid))
previousColors["-1"] = "grey0";
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot = genRainbowHapPlotObj(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates %>%
mutate(popid= factor(popid)), colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name)),
#labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name),
labels = rep("", length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name))),
expand = c(0,0))+
scale_y_continuous(breaks = 1:length(levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$s_Sample)),
labels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$s_Sample),
expand = c(0,0))
```
The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal
```{r}
#| fig-column: screen
#| fig-width: 30
#| fig-height: 5
regions_homologousRegion_filt = regions_homologousRegion %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates$p_name)))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot_final = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot +
scale_fill_manual("Microhaplotype\nRank", values = haplotypeRankColors[sort(names(previousColors))], labels = names(haplotypeRankColors[sort(names(previousColors))]), breaks = names(haplotypeRankColors[sort(names(previousColors))])) +
guides(fill = guide_legend(nrow = 1)) +
ggnewscale::new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -1,
fill = description),
data = regions_homologousRegion_filt, color = "black") +
scale_fill_manual(values = descriptionColors_homologousRegion,
guide = guide_legend(nrow = 2)) +
labs(fill = "Genes\nDescription") +
transparentBackground + theme(legend.text = element_text(size = 30),
legend.title = element_text(size = 30, face = "bold"),
legend.box="vertical", legend.margin=margin(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"),
axis.text.x = element_text(size = 30),
axis.text.y = element_text(size = 30))
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot_final)
```
```{r}
pdf("popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot.pdf", useDingbats = F, width = 30, height = 10)
print(popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_plot_final)
dev.off()
```
### Getting regions with SD01 Multi In Shared
Outputting the regions within the shared region where SD01 has multiple variants
```{r}
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sd01 = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates %>%
filter(s_Sample == "SD01") %>%
group_by(s_Sample, p_name) %>%
mutate(s_COI = length(unique(h_popUID)))
popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sd01_multi = popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sd01 %>%
filter(s_COI > 1)
regions_withSD01MultiInShared = regions %>%
filter(genomicID %in% popClustering_filt_hrp3_pat1_regions_homologousRegion_prep_LabIsolates_sd01_multi$p_name)
write_tsv(regions_withSD01MultiInShared, "regions_withSD01MultiInShared.bed")
finalHrpSubwindows_regions_withSD01MultiInShared = finalHrpSubwindows %>%
filter(X4 %in% regions_withSD01MultiInShared$name)
write_tsv(finalHrpSubwindows_regions_withSD01MultiInShared, "finalHrpSubwindows_regions_withSD01MultiInShared.bed", col_names = F)
```
### Plottng Shared Region for Pacbio Genomes
```{r}
popClustering_labIso = popClustering %>%
left_join(meta %>%
rename(s_Sample = BiologicalSample)) %>%
filter(grepl("^Pf", sample))
popClustering_labIso_homologousRegion = popClustering_labIso %>%
filter(p_name %in% regions_homologousRegion$genomicID)
popClustering_labIso_homologousRegion_prep = HaplotypeRainbows::prepForRainbow(popClustering_labIso_homologousRegion, minPopSize = 1)
```
```{r}
# select just the major haplotypes and cluster based on the sharing between
popClustering_labIso_homologousRegion_prep_sp = popClustering_labIso_homologousRegion_prep %>%
group_by(p_name) %>%
mutate(sampleCount = length(unique(s_Sample)))%>%
group_by()%>%
filter(sampleCount > 0.9*max(sampleCount)) %>%
group_by(s_Sample, p_name) %>%
# filter(c_AveragedFrac == max(c_AveragedFrac)) %>%
mutate(marker = 1) %>%
group_by() %>%
select(h_popUID, marker, s_Sample) %>%
spread(h_popUID, marker, fill = 0)
popClustering_labIso_homologousRegion_prep_sp_mat = as.matrix(popClustering_labIso_homologousRegion_prep_sp[,2:ncol(popClustering_labIso_homologousRegion_prep_sp)])
rownames(popClustering_labIso_homologousRegion_prep_sp_mat) = popClustering_labIso_homologousRegion_prep_sp$s_Sample
popClustering_labIso_homologousRegion_prep_sp_dist = dist(popClustering_labIso_homologousRegion_prep_sp_mat)
popClustering_labIso_homologousRegion_prep_sp_dist_hclust = hclust(popClustering_labIso_homologousRegion_prep_sp_dist)
#rename the levels so they are in the order of the clustering
popClustering_labIso_homologousRegion_prep = popClustering_labIso_homologousRegion_prep %>%
mutate(s_Sample = factor(s_Sample,
levels = rownames(popClustering_labIso_homologousRegion_prep_sp_mat)[popClustering_labIso_homologousRegion_prep_sp_dist_hclust$order]))
popClustering_labIso_homologousRegion_prep_plot = genRainbowHapPlotObj(popClustering_labIso_homologousRegion_prep, colorCol = popid) +
theme(axis.text.x = element_text(size=12, angle = -90, vjust = 0.5, hjust = 0)) +
scale_x_continuous(breaks = 1:length(levels(popClustering_labIso_homologousRegion_prep$p_name)),
labels = levels(popClustering_labIso_homologousRegion_prep$p_name),
expand = c(0,0))+
scale_y_continuous(expand = c(0,0))
```
The y axis is samples and the x-axis is sub region within the chromosome, sorted by genomic position. Haplotypes are colored by their abundance rank and while colors in a vertical column are the same haplotype, the same colors between column do not mean same haplotype, Rank is ordered by frequency within the total population. Columns with black bars are columns where there is no haplotype variation. Bar heights are relative to abundance within sample, so a sample with just 1 bar for a genomic position means monoclonal at this position while multiple bars would indicated polyclonal
```{r}
#| fig-column: screen
#| fig-width: 30
#| fig-height: 5
regions_homologousRegion_filt = regions_homologousRegion %>%
filter(genomicID %in% popClustering_labIso_homologousRegion_prep$p_name) %>%
mutate(genomicID = factor(genomicID, levels = levels(popClustering_labIso_homologousRegion_prep$p_name)))
print(popClustering_labIso_homologousRegion_prep_plot +
new_scale_fill() +
geom_rect(aes(xmin = as.numeric(genomicID) - 0.5,
xmax = as.numeric(genomicID) + 0.5,
ymax = 0,
ymin = -1,
fill = description),
data = regions_homologousRegion_filt, color = "black") +
scale_fill_manual(values = descriptionColors_homologousRegion,
guide = guide_legend(nrow = 2))
)
```
# Plotting whole genome inter-relatednesss between strains with deletions
```{r, echo=T, eval=T}
allSel = readr::read_tsv("/Users/nick/Dropbox (Personal)/ownCloud/documents/plasmodium/falciparum/pfepipanels/Pf_Epi_Panels/data/MIPSIBC/data/pf/reports/slim_allSelectedClustersInfo.tab.txt.gz")
allSel_filt = allSel %>%
filter(s_Sample %in% previousDeletionCalls$BiologicalSample)
write_tsv(allSel_filt, "MIPSIBC_previousDeletionCalls_samples.tsv")
```
```{r, echo=T, eval=T}
allSel = readr::read_tsv("/Users/nick/Dropbox (Personal)/ownCloud/documents/plasmodium/falciparum/pfepipanels/Pf_Epi_Panels/data/heome1/data/pf/reports/slim_allSelectedClustersInfo.tab.txt.gz")
allSel_filt = allSel %>%
filter(s_Sample %in% previousDeletionCalls$BiologicalSample)
write_tsv(allSel_filt, "heome1_previousDeletionCalls_samples.tsv")
```
```{bash, echo=T, eval=F}
elucidator doPairwiseComparisonOnHapsSharing --tableFnp heome1_previousDeletionCalls_samples.tsv --sampleCol s_Sample --targetNameCol p_name --popIDCol h_popUID --relAbundCol c_AveragedFrac --numThreads 12 --dout heome1_previousDeletionCalls_samples_pairwiseComps --verbose --overWriteDir --metaFnp /tank/data/plasmodium/falciparum/pfdata/metadata/metaByBiosample.tab.txt --metaFieldsToCalcPopDiffs country,region,secondaryRegion --writeOutDistMatrices
elucidator doPairwiseComparisonOnHapsSharing --tableFnp MIPSIBC_previousDeletionCalls_samples.tsv --sampleCol s_Sample --targetNameCol p_name --popIDCol h_popUID --relAbundCol c_AveragedFrac --numThreads 12 --dout MIPSIBC_previousDeletionCalls_samples_pairwiseComps --verbose --overWriteDir --metaFnp /tank/data/plasmodium/falciparum/pfdata/metadata/metaByBiosample.tab.txt --metaFieldsToCalcPopDiffs country,region,secondaryRegion --writeOutDistMatrices
```
## Heome1
```{r, echo=T, eval=T}
sample_metadata_withAllDeletionCalls=readr::read_tsv("sample_metadata_withAllDeletionCalls.tsv")
heome1_previousDeletionCalls_samples_jacardByHapsTarShared_samples = readr::read_tsv("heome1_previousDeletionCalls_samples_pairwiseComps/sampleNames.tab.txt", col_names = F)
heome1_previousDeletionCalls_samples_jacardByHapsTarShared = readr::read_tsv("heome1_previousDeletionCalls_samples_pairwiseComps/jacardByHapsTarShared.tab.txt.gz", col_names = F)
heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat = as.matrix(heome1_previousDeletionCalls_samples_jacardByHapsTarShared)
colnames(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat) = heome1_previousDeletionCalls_samples_jacardByHapsTarShared_samples$X1
rownames(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat) = heome1_previousDeletionCalls_samples_jacardByHapsTarShared_samples$X1
heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat = heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat[metaSelected$BiologicalSample,metaSelected$BiologicalSample]
col_fun = colorRamp2(c(min(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat), min(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat) + (1-min(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
previousDeletionCalls_sel = previousDeletionCalls[match(colnames(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat), previousDeletionCalls$BiologicalSample),]%>%
left_join(sample_metadata_withAllDeletionCalls %>%
select(sample, Pattern) %>%
rename(BiologicalSample = sample)) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
mutate(newClusterName = ifelse(hcclustSize == 1, "singlet", as.character(stringr::str_pad(newClusterName, width = 2, pad = "0")))) %>%
mutate(BiologicalSample = as.character(BiologicalSample)) %>%
select(BiologicalSample, newClusterName))
rowAnnoDf = previousDeletionCalls_sel[, c(
"country",
"region",
"secondaryRegion",
"newClusterName",
"Pattern")] %>%
rename(continent = secondaryRegion,
Chr11DupHapCluster = newClusterName) %>%
as.data.frame()
rowAnnoColors = createColorListFromDf(rowAnnoDf)
load("rowAnnoColors.Rdata")
rowAnnoColors_mod = rowAnnoColors
rowAnnoColors_mod[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
rowAnnoColors_mod[["Pattern"]] = rowAnnoColorsMod_hrp3DeletionPattern
annotationTextSize = 25 ;annotationTitleTextSize = 20;
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
show_legend = F,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm = Heatmap(
heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat,
col = col_fun,
name = "JacardIndex",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
na_col = "#FFFFFF00",
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "JacardIndex"
)
)
)
```
```{r}
#| fig-column: screen
#| fig-width: 25
#| fig-height: 15
draw(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
```
```{r}
pdf("heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm.pdf", width = 25, height = 30, useDingbats = F)
draw(heome1_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
dev.off()
```
## MIPSIBC
```{r, echo=T, eval=T}
sample_metadata_withAllDeletionCalls=readr::read_tsv("sample_metadata_withAllDeletionCalls.tsv")
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_samples = readr::read_tsv("MIPSIBC_previousDeletionCalls_samples_pairwiseComps/sampleNames.tab.txt", col_names = F)
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared = readr::read_tsv("MIPSIBC_previousDeletionCalls_samples_pairwiseComps/jacardByHapsTarShared.tab.txt.gz", col_names = F)
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat = as.matrix(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared)
colnames(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat) = MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_samples$X1
rownames(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat) = MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_samples$X1
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat = MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat[metaSelected$BiologicalSample,metaSelected$BiologicalSample]
col_fun = colorRamp2(c(min(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat), min(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat) + (1-min(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
previousDeletionCalls_sel = previousDeletionCalls[match(colnames(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat), previousDeletionCalls$BiologicalSample),]%>%
left_join(sample_metadata_withAllDeletionCalls %>%
select(sample, Pattern) %>%
rename(BiologicalSample = sample)) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
mutate(newClusterName = ifelse(hcclustSize == 1, "singlet", as.character(stringr::str_pad(newClusterName, width = 2, pad = "0")))) %>%
mutate(BiologicalSample = as.character(BiologicalSample)) %>%
select(BiologicalSample, newClusterName))
rowAnnoDf = previousDeletionCalls_sel[, c(
"country",
"region",
"secondaryRegion",
"newClusterName",
"Pattern")] %>%
rename(continent = secondaryRegion,
Chr11DupHapCluster = newClusterName) %>%
as.data.frame()
rowAnnoColors = createColorListFromDf(rowAnnoDf)
load("rowAnnoColors.Rdata")
rowAnnoColors_mod = rowAnnoColors
rowAnnoColors_mod[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
rowAnnoColors_mod[["Pattern"]] = rowAnnoColorsMod_hrp3DeletionPattern
annotationTextSize = 25 ;annotationTitleTextSize = 20;
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
show_legend = F,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_nolabs = MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat
rownames(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_nolabs) = NULL
colnames(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_nolabs) = NULL
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm = Heatmap(
MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_nolabs,
col = col_fun,
name = "JacardIndex",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
na_col = "#FFFFFF00",
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "JacardIndex"
)
)
)
```
```{r}
#| fig-column: screen
#| fig-width: 25
#| fig-height: 15
draw(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
```
```{r}
pdf("MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm.pdf", width = 25, height = 30, useDingbats = F)
draw(MIPSIBC_previousDeletionCalls_samples_jacardByHapsTarShared_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
dev.off()
```
## hmmIBD
```{r}
sample_metadata_withAllDeletionCalls_sel = sample_metadata_withAllDeletionCalls %>%
select(sample, SRARuns) %>%
filter(sample %in% previousDeletionCalls$BiologicalSample) %>%
mutate(SRARuns = strsplit(SRARuns, split = ",")) %>%
unnest(SRARuns)
hmm_fract = readr::read_tsv("filtered_gatk_calls_database_hrpsallMetaDeletionCalls.hmm_fract.txt")
hmm_fract_combined = bind_rows(
hmm_fract %>%
arrange(sample1, sample2) %>%
select(sample1, sample2, fract_sites_IBD),
hmm_fract %>%
arrange(sample1, sample2) %>%
select(sample1, sample2, fract_sites_IBD) %>%
rename(temp1 = sample2,
temp2 = sample1)%>%
rename(sample1 = temp1,
sample2 = temp2)
)
hmm_fract_combined_samples = tibble(sample1 = unique(hmm_fract_combined$sample1)) %>%
left_join(sample_metadata_withAllDeletionCalls_sel %>%
rename(BiologicalSample = sample,
sample1 = SRARuns)) %>%
mutate(BiologicalSample = ifelse(is.na(BiologicalSample), sample1, BiologicalSample)) %>%
mutate(BiologicalSample = ifelse(BiologicalSample == "fcr3", "FCR3", BiologicalSample))
perSampleVarCounts = readr::read_tsv("filtered_hrpsallMetaDeletionCalls_variants_perSampleCounts.tsv") %>%
rename(sample1 = `[3]sample`,
nMissing = `[14]nMissing`)
hmm_fract_combined_samples_filt = hmm_fract_combined_samples %>%
left_join(perSampleVarCounts %>%
select(sample1, nMissing)) %>%
group_by(BiologicalSample) %>%
arrange(BiologicalSample, nMissing) %>%
mutate(rank = row_number(),
totalSamples = n()) %>%
filter(rank == 1)
hmm_fract_combined_filt = hmm_fract_combined %>%
filter(sample1 %in% hmm_fract_combined_samples_filt$sample1,
sample2 %in% hmm_fract_combined_samples_filt$sample1) %>%
left_join(hmm_fract_combined_samples_filt %>%
rename(BiologicalSample1 = BiologicalSample) %>%
select(sample1, BiologicalSample1))%>%
left_join(hmm_fract_combined_samples_filt %>%
rename(
sample2 = sample1,
BiologicalSample2 = BiologicalSample) %>%
select(sample2, BiologicalSample2))
hmm_fract_sp = hmm_fract_combined_filt %>%
select(BiologicalSample1, BiologicalSample2, fract_sites_IBD) %>%
spread(BiologicalSample2, fract_sites_IBD, fill = 1)
hmm_fract_sp_mat = as.matrix(hmm_fract_sp[,2:ncol(hmm_fract_sp)])
rownames(hmm_fract_sp_mat) = hmm_fract_sp$BiologicalSample1
```
```{r}
library(circlize)
hmm_fract_sp_mat = hmm_fract_sp_mat[metaSelected$BiologicalSample, metaSelected$BiologicalSample]
# col_fun = colorRamp2(c(min(hmm_fract_sp_mat), min(hmm_fract_sp_mat) + (1-min(hmm_fract_sp_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
col_fun = colorRamp2(c(0, 0.5, 1), c( "#2166ac", "white", "#b2182b"))
previousDeletionCalls_sel = previousDeletionCalls[match(colnames(hmm_fract_sp_mat), previousDeletionCalls$BiologicalSample),]%>%
left_join(sample_metadata_withAllDeletionCalls %>%
select(sample, Pattern) %>%
rename(BiologicalSample = sample)) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
mutate(newClusterName = ifelse(hcclustSize == 1, "singlet", as.character(stringr::str_pad(newClusterName, width = 2, pad = "0")))) %>%
mutate(BiologicalSample = as.character(BiologicalSample)) %>%
select(BiologicalSample, newClusterName)) %>%
mutate(Pattern = ifelse(is.na(Pattern) & possiblyHRP2Deleted, "8-TARE1", Pattern)) %>%
mutate(Pattern = ifelse("FCR3" == BiologicalSample, "13++11-", Pattern))
rowAnnoDf = previousDeletionCalls_sel[, c(
"country",
"region",
"secondaryRegion",
"newClusterName",
"Pattern",
"hrpCall")] %>%
rename(continent = secondaryRegion,
Chr11DupHapCluster = newClusterName) %>%
as.data.frame()
rowAnnoColors = createColorListFromDf(rowAnnoDf)
load("rowAnnoColors.Rdata")
rowAnnoColors_mod = rowAnnoColors
rowAnnoColors_mod[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
rowAnnoColors_mod[["Pattern"]] = rowAnnoColorsMod_hrp3DeletionPattern
annotationTextSize = 25 ;annotationTitleTextSize = 20;
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
show_legend = F,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
hmm_fract_sp_mat_nolabs = hmm_fract_sp_mat
rownames(hmm_fract_sp_mat_nolabs) = NULL
colnames(hmm_fract_sp_mat_nolabs) = NULL
hmm_fract_sp_mat_hm = Heatmap(
hmm_fract_sp_mat_nolabs,
col = col_fun,
name = "fracIBDSites",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
na_col = "#FFFFFF00" ,
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "fracIBDSites"
)
)
)
```
```{r}
#| fig-column: screen
#| fig-width: 27
#| fig-height: 30
draw(hmm_fract_sp_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
```
```{r}
pdf("hmmIBD_fract_sp_mat_hm.pdf", width = 25, height = 30, useDingbats = F)
draw(hmm_fract_sp_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
dev.off()
```
### hmmIBD for just 13-11++ parasites
```{r}
hmm_fract_sp_pat1 = hmm_fract_combined_filt %>%
filter(BiologicalSample1 %in% metaSelected_hrp3_pat1$BiologicalSample,
BiologicalSample2 %in% metaSelected_hrp3_pat1$BiologicalSample) %>%
select(BiologicalSample1, BiologicalSample2, fract_sites_IBD) %>%
spread(BiologicalSample2, fract_sites_IBD, fill = 1)
hmm_fract_sp_pat1_mat = as.matrix(hmm_fract_sp_pat1[,2:ncol(hmm_fract_sp_pat1)])
rownames(hmm_fract_sp_pat1_mat) = hmm_fract_sp_pat1$BiologicalSample1
```
```{r}
library(circlize)
# col_fun = colorRamp2(c(min(hmm_fract_sp_pat1_mat), min(hmm_fract_sp_pat1_mat) + (1-min(hmm_fract_sp_pat1_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
col_fun = colorRamp2(c(0, 0.5, 1), c( "#2166ac", "white", "#b2182b"))
previousDeletionCalls_sel = previousDeletionCalls[match(colnames(hmm_fract_sp_pat1_mat), previousDeletionCalls$BiologicalSample),]%>%
left_join(sample_metadata_withAllDeletionCalls %>%
select(sample, Pattern) %>%
rename(BiologicalSample = sample)) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
mutate(newClusterName = ifelse(hcclustSize == 1, "singlet", as.character(stringr::str_pad(newClusterName, width = 2, pad = "0")))) %>%
mutate(BiologicalSample = as.character(BiologicalSample)) %>%
select(BiologicalSample, newClusterName))
rowAnnoDf = previousDeletionCalls_sel[, c(
"country",
"region",
"secondaryRegion",
"newClusterName",
# "Pattern",
"hrpCall")] %>%
rename(continent = secondaryRegion,
Chr11DupHapCluster = newClusterName) %>%
as.data.frame()
rowAnnoColors = createColorListFromDf(rowAnnoDf)
load("rowAnnoColors.Rdata")
rowAnnoColors_mod = rowAnnoColors
rowAnnoColors_mod[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
rowAnnoColors_mod[["Pattern"]] = rowAnnoColorsMod_hrp3DeletionPattern
annotationTextSize = 25 ;annotationTitleTextSize = 20;
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
show_legend = F,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
hmm_fract_sp_pat1_mat_nolabs = hmm_fract_sp_pat1_mat
rownames(hmm_fract_sp_pat1_mat_nolabs) = NULL
colnames(hmm_fract_sp_pat1_mat_nolabs) = NULL
hmm_fract_sp_pat1_mat_hm = Heatmap(
hmm_fract_sp_pat1_mat_nolabs,
col = col_fun,
name = "fracIBDSites",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
na_col = "#FFFFFF00" ,
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "fracIBDSites"
)
)
)
```
```{r}
#| fig-column: screen
#| fig-width: 27
#| fig-height: 30
draw(hmm_fract_sp_pat1_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
```
```{r}
pdf("hmmIBD_fract_sp_mat_hm_pat1.pdf", width = 25, height = 25, useDingbats = F)
draw(hmm_fract_sp_pat1_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
dev.off()
```
### hmmIBD for just 13-5++ parasites
```{r}
sample_metadata_withAllDeletionCalls_13_5_pattern = sample_metadata_withAllDeletionCalls %>%
filter(Pattern == "13-5++")
hmm_fract_sp_13_5_pattern = hmm_fract_combined_filt %>%
filter(BiologicalSample1 %in% sample_metadata_withAllDeletionCalls_13_5_pattern$sample,
BiologicalSample2 %in% sample_metadata_withAllDeletionCalls_13_5_pattern$sample) %>%
select(BiologicalSample1, BiologicalSample2, fract_sites_IBD) %>%
spread(BiologicalSample2, fract_sites_IBD, fill = 1)
hmm_fract_sp_13_5_pattern_mat = as.matrix(hmm_fract_sp_13_5_pattern[,2:ncol(hmm_fract_sp_13_5_pattern)])
rownames(hmm_fract_sp_13_5_pattern_mat) = hmm_fract_sp_13_5_pattern$BiologicalSample1
```
```{r}
library(circlize)
# col_fun = colorRamp2(c(min(hmm_fract_sp_13_5_pattern_mat), min(hmm_fract_sp_13_5_pattern_mat) + (1-min(hmm_fract_sp_13_5_pattern_mat))/2, 1), c( "#2166ac", "white", "#b2182b"))
col_fun = colorRamp2(c(0, 0.5, 1), c( "#2166ac", "white", "#b2182b"))
previousDeletionCalls_sel = previousDeletionCalls[match(colnames(hmm_fract_sp_13_5_pattern_mat), previousDeletionCalls$BiologicalSample),]%>%
left_join(sample_metadata_withAllDeletionCalls %>%
select(sample, Pattern) %>%
rename(BiologicalSample = sample)) %>%
left_join(popClustering_filt_hrp3_pat1_regions_afterHomologous_chr11_prep_forClustering_sp_dist_hclust_groups_df %>%
ungroup() %>%
mutate(newClusterName = ifelse(hcclustSize == 1, "singlet", as.character(stringr::str_pad(newClusterName, width = 2, pad = "0")))) %>%
mutate(BiologicalSample = as.character(BiologicalSample)) %>%
select(BiologicalSample, newClusterName))
rowAnnoDf = previousDeletionCalls_sel[, c(
"country",
"region",
"secondaryRegion"
# "newClusterName",
# "Pattern",
# "hrpCall"
)] %>%
rename(continent = secondaryRegion) %>%
as.data.frame()
rowAnnoColors = createColorListFromDf(rowAnnoDf)
load("rowAnnoColors.Rdata")
rowAnnoColors_mod = rowAnnoColors
# rowAnnoColors_mod[["Chr11DupHapCluster"]] = newHaploGroupWithSingletColors
rowAnnoColors_mod[["Pattern"]] = rowAnnoColorsMod_hrp3DeletionPattern
annotationTextSize = 25 ;annotationTitleTextSize = 20;
topAnno = HeatmapAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
show_legend = F,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
sideAnno = rowAnnotation(
df = rowAnnoDf,
col = rowAnnoColors_mod,
gp = gpar(col = "grey10"),
annotation_name_gp = gpar(fontsize = annotationTitleTextSize),
annotation_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(fontsize = annotationTextSize, fontface = "bold")
),
na_col = "#FFFFFF00"
)
hmm_fract_sp_13_5_pattern_mat_nolabs = hmm_fract_sp_13_5_pattern_mat
rownames(hmm_fract_sp_13_5_pattern_mat_nolabs) = NULL
colnames(hmm_fract_sp_13_5_pattern_mat_nolabs) = NULL
hmm_fract_sp_13_5_pattern_mat_hm = Heatmap(
hmm_fract_sp_13_5_pattern_mat_nolabs,
col = col_fun,
name = "fracIBDSites",
top_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
na_col = "#FFFFFF00" ,
heatmap_legend_param = list(
labels_gp = gpar(fontsize = annotationTextSize),
title_gp = gpar(
fontsize = annotationTextSize,
fontface = "bold",
title = "fracIBDSites"
)
)
)
```
```{r}
#| fig-column: screen
#| fig-width: 15
#| fig-height: 15
draw(hmm_fract_sp_13_5_pattern_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
```
```{r}
pdf("hmmIBD_fract_sp_mat_hm_13_5_pattern.pdf", width = 15, height = 15, useDingbats = F)
draw(hmm_fract_sp_13_5_pattern_mat_hm, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "bottom")
dev.off()
```
## moire
```{r, echo=T, eval = F}
remotes::install_github("EPPIcenter/moire")
setwd("/tank/data/plasmodium/falciparum/pfdata/moire_on_hrp3Samples/")
df <- read.tsv("allSel_withDeletions_prep_outForMoire.tsv")
data <- load_long_form_data(df)
# With data in appropriate format, run MCMC as follows
mcmc_results <- moire::run_mcmc(data, is_missing = data$is_missing)
write_rds(mcmc_results, "mcmc_results.rds")
write_rds(data, "data_for_moire.rds")
```
```{r}
data_for_moire = read_rds("moire_on_hrp3Samples/data_for_moire.rds")
mcmc_results = read_rds("moire_on_hrp3Samples/mcmc_results.rds")
coiEsts = tibble(
sampleID = data_for_moire$sample_ids,
medianCOI = round(unlist(lapply(mcmc_results$chains[[1]]$coi, median))),
meanCOI = round(unlist(lapply(mcmc_results$chains[[1]]$coi, mean))),
maxCOI = round(unlist(lapply(mcmc_results$chains[[1]]$coi, max)))
)
```