Code
genomes = readr::read_tsv("assembly_study_links.txt")
create_dt(genomes)Comparing to genome assembly of all other six Plasmodium species within the subgenus Laverania(Otto et al. 2018)
Theoretical timeline of the P. Laverania subgenus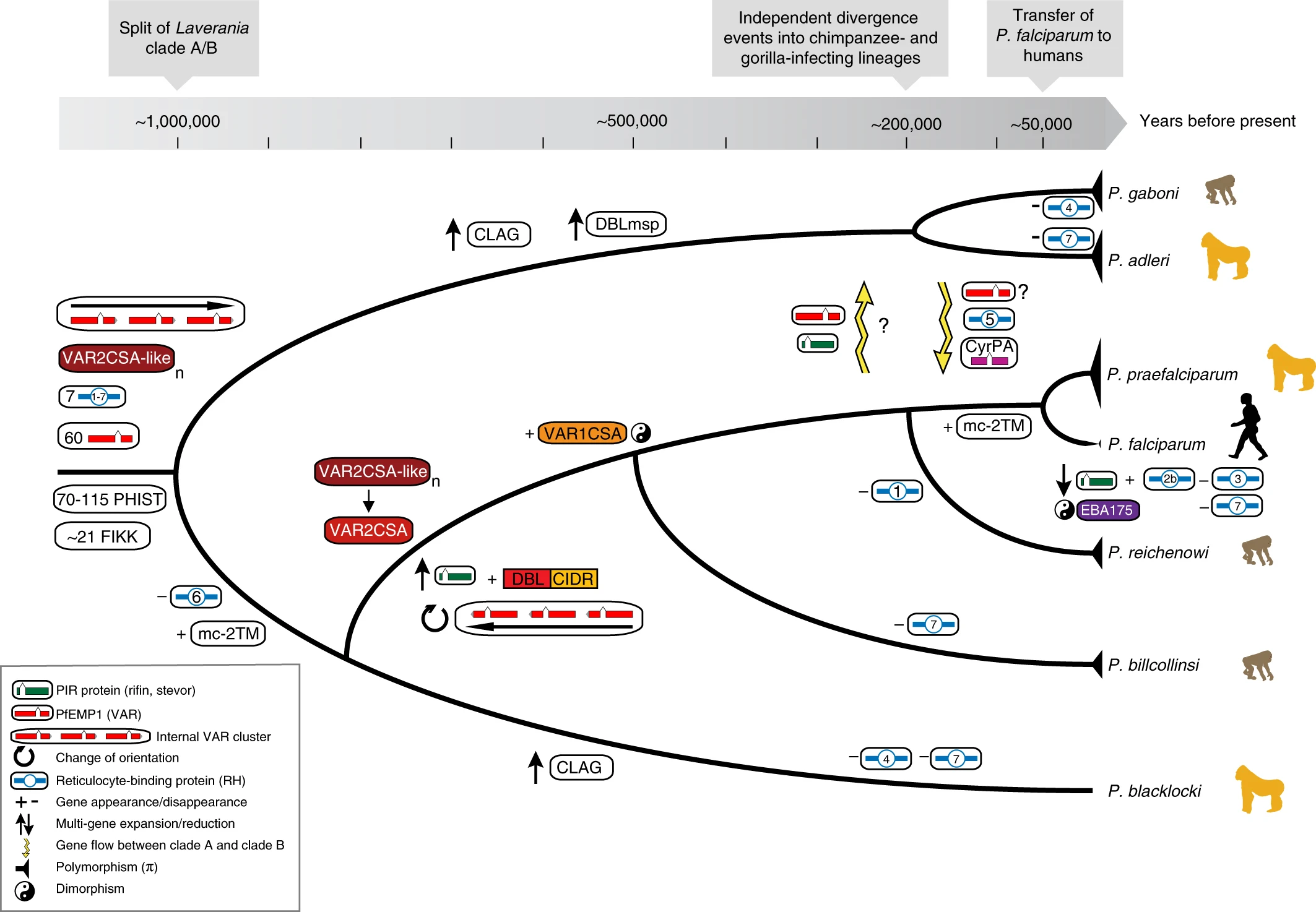
genomes = readr::read_tsv("assembly_study_links.txt")
create_dt(genomes)/tank/data/plasmodium/genomes/pLaverania/genomes/PadleriG01.fasta
/tank/data/plasmodium/genomes/pLaverania/genomes/PbillcollinsiG01.fasta
/tank/data/plasmodium/genomes/pLaverania/genomes/PblacklockiG01.fasta
/tank/data/plasmodium/genomes/pLaverania/genomes/PgaboniG01.fasta
/tank/data/plasmodium/genomes/pLaverania/genomes/PpraefalciparumG01.fasta
/tank/data/plasmodium/genomes/pLaverania/genomes/PreichenowiG01.fasta
runTRFCmds.txt
runNucmerCmds.txt
CMD:nucmer /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.fasta /tank/data/genomes/plasmodium/genomes/pLaverania/genomes/{SPECIES}.fasta --prefix {SPECIES}_to_Pf3D7_nucmer > nuclog_{SPECIES}.txt 2>&1 && show-coords -T -l -c -H {SPECIES}_to_Pf3D7_nucmer.delta | elucidator parseNucmerResultsToBed --coordsOutput STDIN --overWrite --out {SPECIES}_to_Pf3D7_nucmer.delta.bed && elucidator splitColumnContainingMeta --file {SPECIES}_to_Pf3D7_nucmer.delta.bed --delim tab --column col.6 --removeEmptyColumn --addHeader --overWrite --out {SPECIES}_to_Pf3D7_nucmer.delta.tsv && elucidator filterBedRecordsByLength --bed {SPECIES}_to_Pf3D7_nucmer.delta.bed --minLen 2000 --overWrite --out minlen2000_{SPECIES}_to_Pf3D7_nucmer.delta.bed && elucidator bedGetIntersectingGenesInGff --gff /tank/data/genomes/plasmodium/genomes/pf/info/gff/Pf3D7.gff --extraAttributes description --bed minlen2000_{SPECIES}_to_Pf3D7_nucmer.delta.bed --out minlen2000_{SPECIES}_to_Pf3D7_nucmer.delta_withGeneInfo.bed
{SPECIES}:species.txt ---
title: "*Plasmodium Laverania*"
---
```{r setup, echo=FALSE, message=FALSE}
source("../common.R")
```
Comparing to genome assembly of all other six *Plasmodium* species within the subgenus Laverania[@Otto2018-mb]
Theoretical timeline of the *P. Laverania* subgenus


```{r}
genomes = readr::read_tsv("assembly_study_links.txt")
create_dt(genomes)
```
/tank/data/plasmodium/genomes/pLaverania/genomes/PadleriG01.fasta
/tank/data/plasmodium/genomes/pLaverania/genomes/PbillcollinsiG01.fasta
/tank/data/plasmodium/genomes/pLaverania/genomes/PblacklockiG01.fasta
/tank/data/plasmodium/genomes/pLaverania/genomes/PgaboniG01.fasta
/tank/data/plasmodium/genomes/pLaverania/genomes/PpraefalciparumG01.fasta
/tank/data/plasmodium/genomes/pLaverania/genomes/PreichenowiG01.fasta
## tandem repeat finder
```{bash, eval = F}
/bin/ls /tank/data/plasmodium/genomes/pLaverania/genomes/*.fasta | sed 's/.*\//g' | sed 's/.fasta/g' > species.txt
```
**runTRFCmds.txt**
```{bash, eval = F}
CMD:elucidator runTRF --fasta /tank/data/plasmodium/genomes/pLaverania/genomes/{SPECIES}.fasta --supplement --dout trf_{SPECIES} --overWriteDir
{SPECIES}:species.txt
```
```{bash, eval = F}
nohup elucidator runMultipleCommands --cmdFile runTRFCmds.txt --numThreads 6 &
```
## nucmer
**runNucmerCmds.txt**
```{bash, eval = F}
CMD:nucmer /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.fasta /tank/data/genomes/plasmodium/genomes/pLaverania/genomes/{SPECIES}.fasta --prefix {SPECIES}_to_Pf3D7_nucmer > nuclog_{SPECIES}.txt 2>&1 && show-coords -T -l -c -H {SPECIES}_to_Pf3D7_nucmer.delta | elucidator parseNucmerResultsToBed --coordsOutput STDIN --overWrite --out {SPECIES}_to_Pf3D7_nucmer.delta.bed && elucidator splitColumnContainingMeta --file {SPECIES}_to_Pf3D7_nucmer.delta.bed --delim tab --column col.6 --removeEmptyColumn --addHeader --overWrite --out {SPECIES}_to_Pf3D7_nucmer.delta.tsv && elucidator filterBedRecordsByLength --bed {SPECIES}_to_Pf3D7_nucmer.delta.bed --minLen 2000 --overWrite --out minlen2000_{SPECIES}_to_Pf3D7_nucmer.delta.bed && elucidator bedGetIntersectingGenesInGff --gff /tank/data/genomes/plasmodium/genomes/pf/info/gff/Pf3D7.gff --extraAttributes description --bed minlen2000_{SPECIES}_to_Pf3D7_nucmer.delta.bed --out minlen2000_{SPECIES}_to_Pf3D7_nucmer.delta_withGeneInfo.bed
{SPECIES}:species.txt
```
```{bash, eval = F}
nohup elucidator runMultipleCommands --cmdFile runNucmerCmds.txt --numThreads 6 &
```