---
title: Comparing To 3D7 Chr11 and 13
---
```{r setup, echo=FALSE, message=FALSE}
source("../../common.R")
```
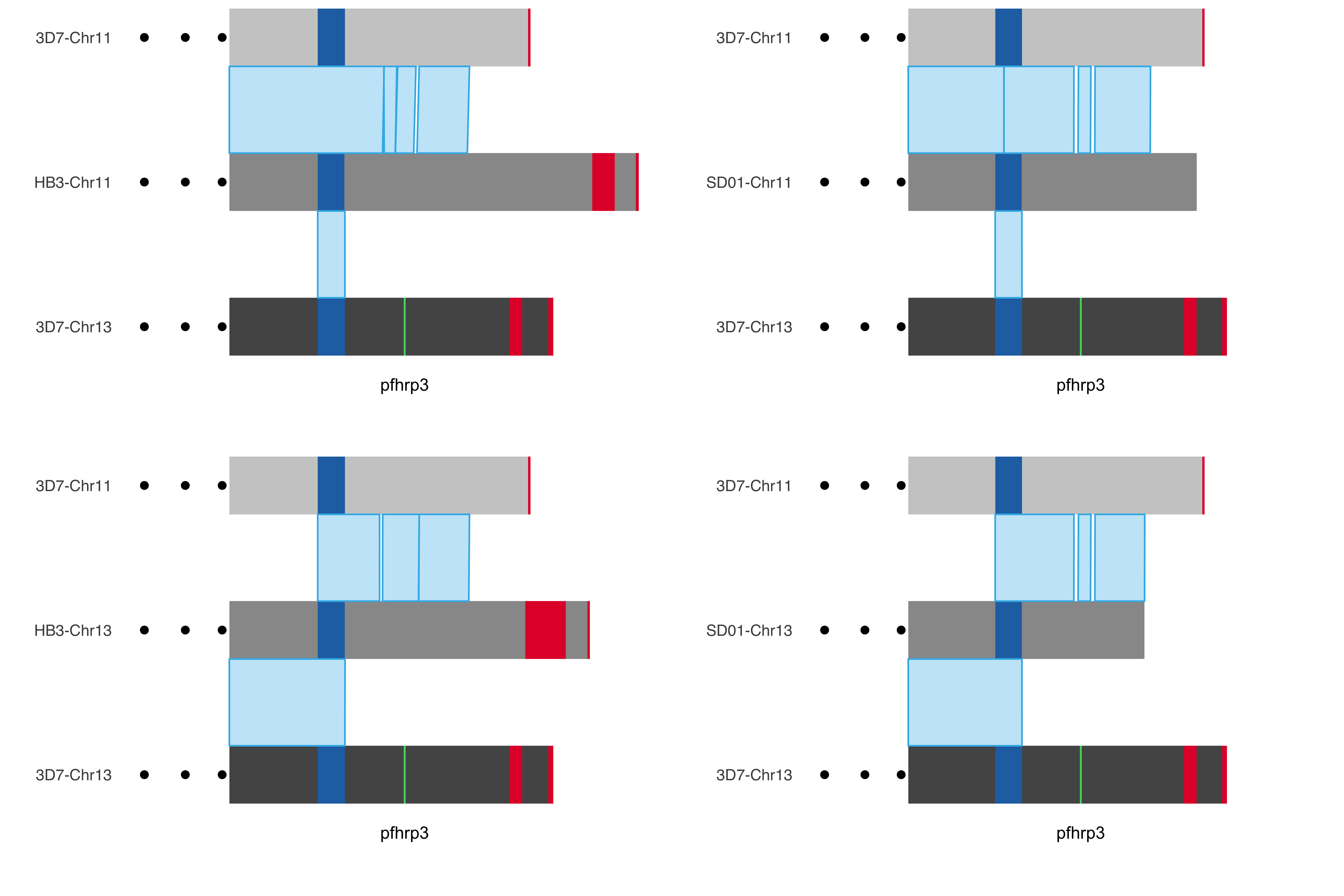
## Comparing SD01 and HB3 assembly to 3D7 chr 11 and 13
```{r}
hb3_repeats = readr::read_tsv("../../HB3_new_assembly/repeats_filt_starts_ends_tares.tsv")
hb3_tares = hb3_repeats %>%
filter(tare1 | SB3)
threed7_repeats = readr::read_tsv("../../MappingOutSurroundingRegions/pf3D7_repeats_filt_starts_ends_tares.tsv")
threed7_tares = threed7_repeats %>%
filter(tare1 | SB3)
HB3_vs_3D7_nucmer = readr::read_tsv("../../HB3_new_assembly/PfHB3nano_to_Pf3D7_nucmer.delta.tsv")
ChromsHB3 = readr::read_tsv("../../HB3_new_assembly/PfHB3nano_chrom_length.tab.txt")
Chroms3D7 = readr::read_tsv("../../HB3_new_assembly/Pf3D7_chrom_length.tab.txt")
SD01_vs_3D7_nucmer = readr::read_tsv("../../SD01_new_assembly/ownAssemblies/combining_flyeAssemblyDefaultForChr11_ForChr13/PfSD01nano_to_Pf3D7_nucmer.delta.tsv")
ChromsSD01 = readr::read_tsv("../../SD01_new_assembly/ownAssemblies/combining_flyeAssemblyDefaultForChr11_ForChr13/chroms_pilon.txt", col_names = c("chrom", "length"))
```
```{bash, eval = F}
cd ../../HB3_new_assembly/
elucidator tableExtractCriteria --file PfHB3nano_to_Pf3D7_nucmer.delta.tsv --delim tab --header --columnName perID --cutOff 92.1 | tail -n +2 > PfHB3nano_to_Pf3D7_nucmer_filt_perIDCutOff92.1.bed
elucidator mergeOverlappingRegionsSameName --bedFnp PfHB3nano_to_Pf3D7_nucmer_filt_perIDCutOff92.1.bed | cut -f1-6 > merged_PfHB3nano_to_Pf3D7_nucmer_filt_perIDCutOff92.1.bed
```
```{bash, eval = F}
cd ../../SD01_new_assembly/ownAssemblies/combining_flyeAssemblyDefaultForChr11_ForChr13
elucidator tableExtractCriteria --file PfSD01nano_to_Pf3D7_nucmer.delta.tsv --delim tab --header --columnName perID --cutOff 92.1 | tail -n +2 > PfSD01nano_to_Pf3D7_nucmer_filt_perIDCutOff92.1.bed
elucidator mergeOverlappingRegionsSameName --bedFnp PfSD01nano_to_Pf3D7_nucmer_filt_perIDCutOff92.1.bed | cut -f1-6 > merged_PfSD01nano_to_Pf3D7_nucmer_filt_perIDCutOff92.1.bed
```
```{r}
sharedRegion_3D7 = readr::read_tsv("../../../sharedBetween11_and_13/investigatingChrom11Chrom13/shared_11_13_region.bed") %>%
rename(chrom = `#chrom`)
sharedRegion_HB3 = readr::read_tsv("../../HB3_new_assembly/sharedRegionBlast/reOriented_PfHB3_nanopore_11_13/regions.bed", col_names = F) %>%
unique() %>%
mutate(X1 = gsub("PfHB3", "PfHB3nano", X1))
sharedRegion_SD01 = readr::read_tsv("../../SD01_new_assembly/ownAssemblies/combining_flyeAssemblyDefaultForChr11_ForChr13/sharedRegionBlast/contig_73_chr13_pilon/regions.bed", col_names = F) %>%
bind_rows(readr::read_tsv("../../SD01_new_assembly/ownAssemblies/combining_flyeAssemblyDefaultForChr11_ForChr13/sharedRegionBlast/contig_74_pilon/regions.bed", col_names = F)) %>%
unique()
hrpIII_3D7 = readr::read_tsv("../../../mappingOutSurroundingRegions/extractedHRPIII/Pf3D7_13_v3-2840726-2841703-rev/beds/Pf3D7_region.bed", col_names = F)
backtrace = 50000
perIDCutOff = 92.1
lengthCutOff = 4000
```
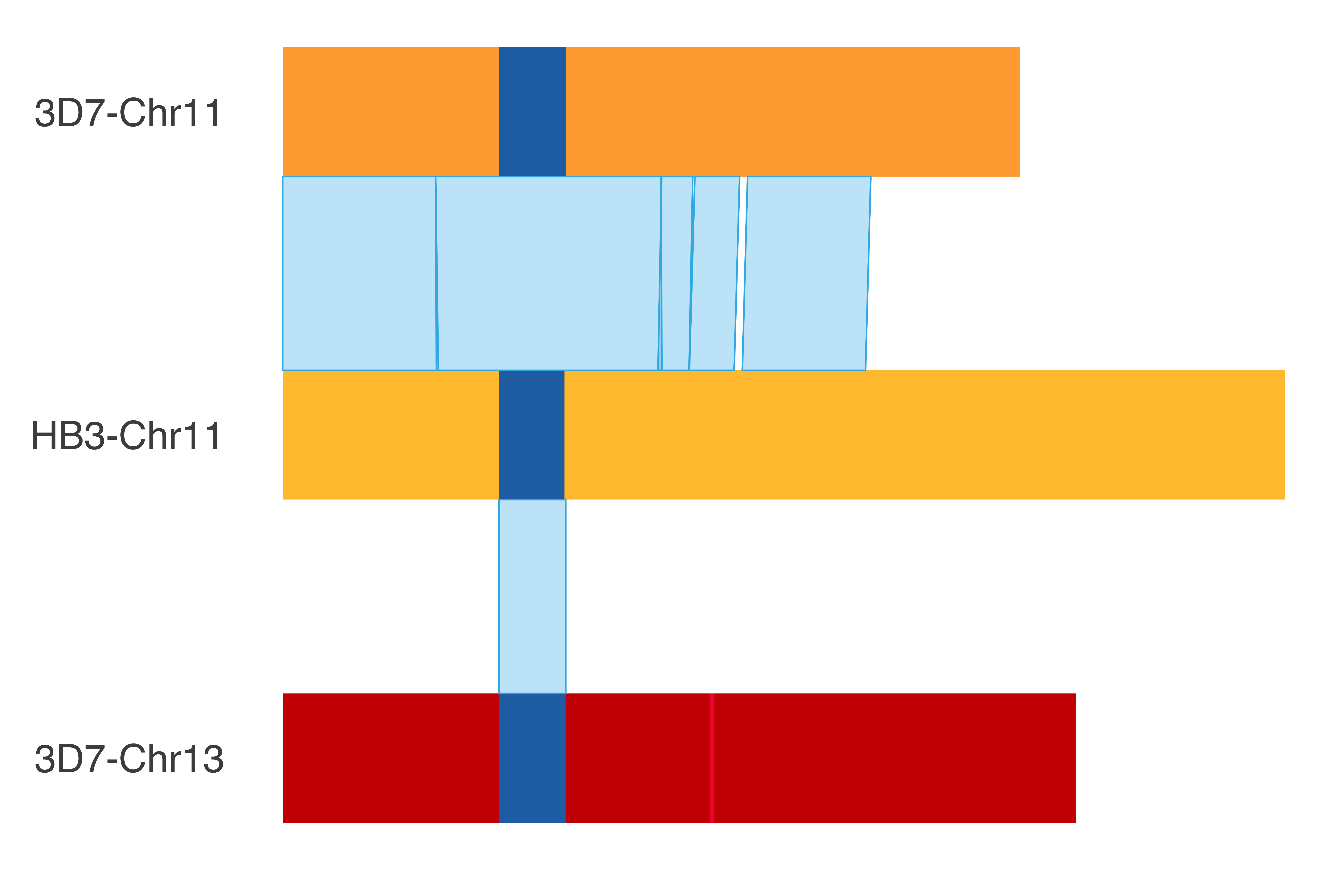
### HB3_chr11
```{r}
HB3_chr11_vs_3D7_chr11_chr13_nucmer = HB3_vs_3D7_nucmer %>%
filter(col.0 %in% c("Pf3D7_11_v3", "Pf3D7_13_v3"),
col.3 %in% c("PfHB3nano_11")) %>%
left_join(sharedRegion_HB3 %>%
select(X1, X2) %>%
filter(X1 == "PfHB3nano_11") %>%
dplyr::rename(HB3SharedStart = X2,
col.3 = X1)) %>%
left_join(sharedRegion_3D7 %>%
select(chrom, start) %>%
rename(col.0 = chrom,
`3D7SharedStart` = start))
HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt = HB3_chr11_vs_3D7_chr11_chr13_nucmer %>%
filter(col.2 >= (`3D7SharedStart` - backtrace))%>%
filter(actualEnd >= (HB3SharedStart - backtrace)) %>%
mutate(`3D7StartAdjust` = col.1 - (`3D7SharedStart` - backtrace),
HB3StartAdjust = actualStart - (HB3SharedStart - backtrace)) %>%
mutate(`3D7EndAdjust` = `3D7StartAdjust` + col.4,
HB3EndAdjust = HB3StartAdjust + col.4) %>%
group_by(col.0, col.1, col.2, col.3, actualStart, actualEnd) %>%
mutate(`3D7StartAdjust` = max(0, `3D7StartAdjust`),
HB3StartAdjust = max(HB3StartAdjust, 0)) %>%
ungroup()
HB3_chr11_chrom = ChromsHB3 %>%
filter(chrom == "PfHB3nano_11") %>%
left_join(HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.3, HB3SharedStart) %>%
rename(chrom = col.3) %>%
unique()) %>%
mutate(start = 0, end = length - (HB3SharedStart - backtrace))
sharedRegion_HB3_chr11_chrom = sharedRegion_HB3 %>%
rename(chrom = X1) %>%
filter(chrom == "PfHB3nano_11") %>%
left_join(HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.3, HB3SharedStart) %>%
rename(chrom = col.3) %>%
unique()) %>%
mutate(start = X2 - (HB3SharedStart - backtrace), end = X3 - (HB3SharedStart - backtrace))
Chroms3D7_chr11 = Chroms3D7 %>%
filter(chrom == "Pf3D7_11_v3") %>%
left_join(HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = 0, end = length - (`3D7SharedStart` - backtrace))
sharedRegion_3D7_chr11_chrom = sharedRegion_3D7 %>%
rename(X2 = start,
X3 = end) %>%
filter(chrom == "Pf3D7_11_v3") %>%
left_join(HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace), end = X3 - (`3D7SharedStart` - backtrace))
Chroms3D7_chr13 = Chroms3D7 %>%
filter(chrom == "Pf3D7_13_v3")%>%
left_join(HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = 0, end = length - (`3D7SharedStart` - backtrace))
sharedRegion_3D7_chr13_chrom = sharedRegion_3D7 %>%
rename(X2 = start,
X3 = end) %>%
filter(chrom == "Pf3D7_13_v3") %>%
left_join(HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace), end = X3 - (`3D7SharedStart` - backtrace))
hrpIII_3D7_chr13_chrom = hrpIII_3D7 %>%
rename(chrom = X1) %>%
filter(chrom == "Pf3D7_13_v3") %>%
left_join(HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace), end = X3 - (`3D7SharedStart` - backtrace))
HB3_chr11_vs_3D7_chr11_nucmer_filt_ribbon = HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff) %>%
mutate(rowid = row_number()) %>%
select(HB3StartAdjust, HB3EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.75) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 1.5)%>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup())
HB3_chr11_vs_3D7_chr13_nucmer_filt_ribbon = HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(HB3StartAdjust, HB3EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.25) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = -0.5)%>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup())
HB3_chr11_chrom_plot = ggplot() +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#FFAA40",
data = Chroms3D7_chr11) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#2370B2",
data = sharedRegion_3D7_chr11_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#CC0200",
data = Chroms3D7_chr13) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1, ymax = -0.5),
fill = "#2370B2",
data = sharedRegion_3D7_chr13_chrom)+
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#F60239",
data = hrpIII_3D7_chr13_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#FFC33B",
data = HB3_chr11_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#2370B2",
data = sharedRegion_HB3_chr11_chrom) +
# geom_rect(aes(xmin = `3D7StartAdjust`, xmax = `3D7StartAdjust` + col.4,
# ymin = 1.5, ymax = 2.0),
# data = HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
# geom_rect(aes(xmin = `3D7StartAdjust`, xmax = `3D7StartAdjust` + col.4,
# ymin = -1.0, ymax = -0.5),
# data = HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
# geom_rect(aes(xmin = `HB3StartAdjust`, xmax = `HB3StartAdjust` + col.4,
# ymin = 0.5, ymax = 0.75),
# data = HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
# geom_rect(aes(xmin = `HB3StartAdjust`, xmax = `HB3StartAdjust` + col.4,
# ymin = 0.25, ymax = 0.5),
# data = HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
geom_polygon(data = HB3_chr11_vs_3D7_chr11_nucmer_filt_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
geom_polygon(data = HB3_chr11_vs_3D7_chr13_nucmer_filt_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
sofonias_theme +
scale_y_continuous(breaks = c(-0.75, 0.5, 1.75),
labels = c("3D7-Chr13", "HB3-Chr11", "3D7-Chr11")) +
labs(y = "", x = "") +
theme(panel.border = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 25))
```
```{r}
#| fig-column: screen-inset
print(HB3_chr11_chrom_plot)
```
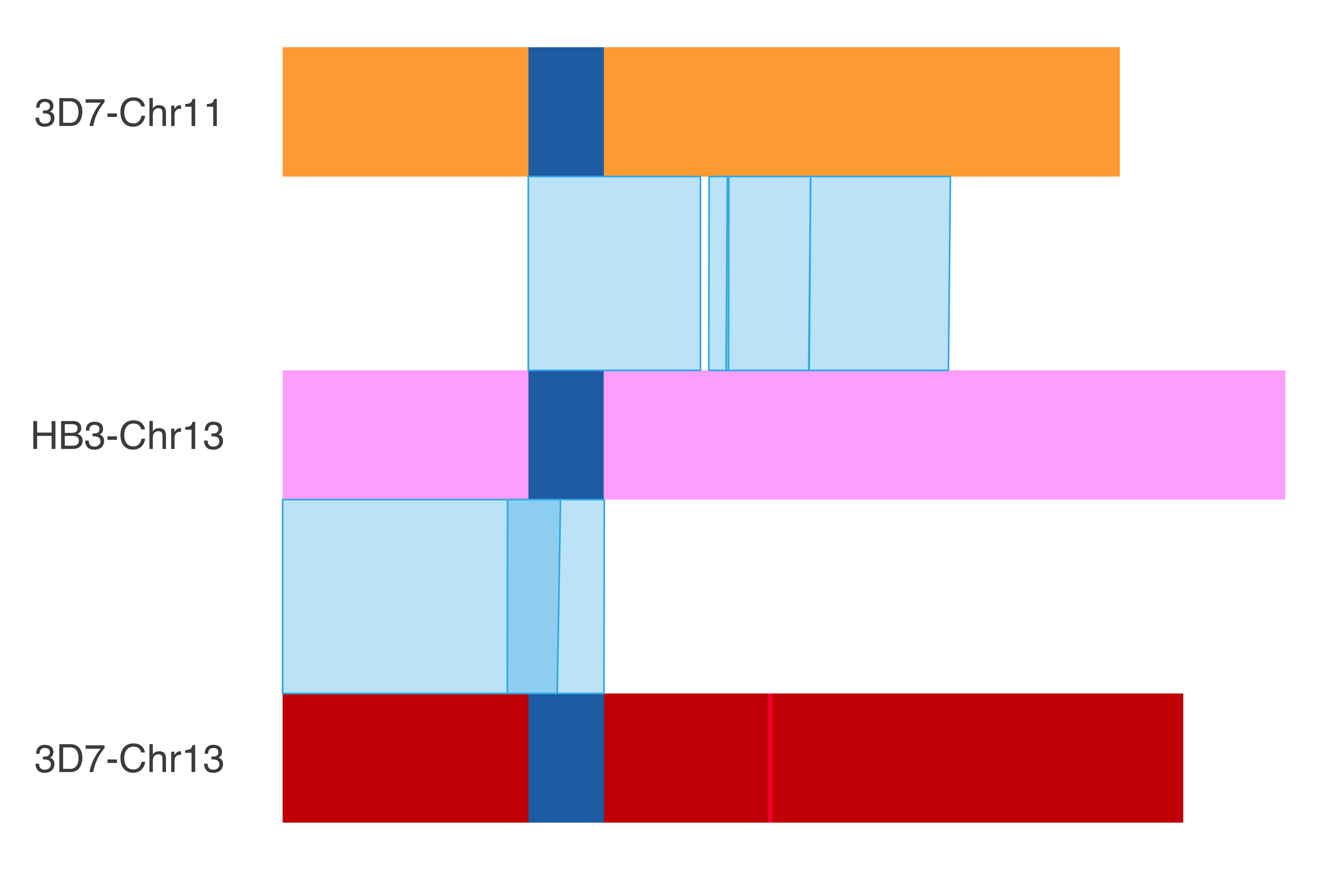
### HB3_chr13
```{r}
HB3_chr13_vs_3D7_chr11_chr13_nucmer = HB3_vs_3D7_nucmer %>%
filter(col.0 %in% c("Pf3D7_11_v3", "Pf3D7_13_v3"),
col.3 %in% c("PfHB3nano_13")) %>%
left_join(sharedRegion_HB3 %>%
select(X1, X2) %>%
filter(X1 == "PfHB3nano_13") %>%
dplyr::rename(HB3SharedStart = X2,
col.3 = X1)) %>%
left_join(sharedRegion_3D7 %>%
select(chrom, start) %>%
rename(col.0 = chrom,
`3D7SharedStart` = start))
HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt = HB3_chr13_vs_3D7_chr11_chr13_nucmer %>%
filter(col.2 >= (`3D7SharedStart` - backtrace))%>%
filter(actualEnd >= (HB3SharedStart - backtrace)) %>%
mutate(`3D7StartAdjust` = col.1 - (`3D7SharedStart` - backtrace),
HB3StartAdjust = actualStart - (HB3SharedStart - backtrace)) %>%
mutate(`3D7EndAdjust` = `3D7StartAdjust` + col.4,
HB3EndAdjust = HB3StartAdjust + col.4) %>%
group_by(col.0, col.1, col.2, col.3, actualStart, actualEnd) %>%
mutate(`3D7StartAdjust` = max(0, `3D7StartAdjust`),
HB3StartAdjust = max(HB3StartAdjust, 0)) %>%
ungroup()
HB3_chr13_chrom = ChromsHB3 %>%
filter(chrom == "PfHB3nano_13") %>%
left_join(HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.3, HB3SharedStart) %>%
rename(chrom = col.3) %>%
unique()) %>%
mutate(start = 0, end = length - (HB3SharedStart - backtrace))
sharedRegion_HB3_chr13_chrom = sharedRegion_HB3 %>%
rename(chrom = X1) %>%
filter(chrom == "PfHB3nano_13") %>%
left_join(HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.3, HB3SharedStart) %>%
rename(chrom = col.3) %>%
unique()) %>%
mutate(start = X2 - (HB3SharedStart - backtrace), end = X3 - (HB3SharedStart - backtrace))
Chroms3D7_chr11 = Chroms3D7 %>%
filter(chrom == "Pf3D7_11_v3") %>%
left_join(HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = 0, end = length - (`3D7SharedStart` - backtrace))
sharedRegion_3D7_chr11_chrom = sharedRegion_3D7 %>%
rename(X2 = start,
X3 = end) %>%
filter(chrom == "Pf3D7_11_v3") %>%
left_join(HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace), end = X3 - (`3D7SharedStart` - backtrace))
Chroms3D7_chr13 = Chroms3D7 %>%
filter(chrom == "Pf3D7_13_v3")%>%
left_join(HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = 0, end = length - (`3D7SharedStart` - backtrace))
sharedRegion_3D7_chr13_chrom = sharedRegion_3D7 %>%
rename(X2 = start,
X3 = end) %>%
filter(chrom == "Pf3D7_13_v3") %>%
left_join(HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace), end = X3 - (`3D7SharedStart` - backtrace))
hrpIII_3D7_chr13_chrom = hrpIII_3D7 %>%
rename(chrom = X1) %>%
filter(chrom == "Pf3D7_13_v3") %>%
left_join(HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace), end = X3 - (`3D7SharedStart` - backtrace))
HB3_chr13_vs_3D7_chr11_nucmer_filt_ribbon = HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(HB3StartAdjust, HB3EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.75) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 1.5)%>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup())
HB3_chr13_vs_3D7_chr13_nucmer_filt_ribbon = HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(HB3StartAdjust, HB3EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.25) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = -0.5)%>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup())
HB3_chr13_chrom_plot = ggplot() +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#FFAA40",
data = Chroms3D7_chr11) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#2370B2",
data = sharedRegion_3D7_chr11_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#CC0200",
data = Chroms3D7_chr13) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1, ymax = -0.5),
fill = "#2370B2",
data = sharedRegion_3D7_chr13_chrom)+
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#F60239",
data = hrpIII_3D7_chr13_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#FFB2FD",
data = HB3_chr13_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#2370B2",
data = sharedRegion_HB3_chr13_chrom) +
# geom_rect(aes(xmin = `3D7StartAdjust`, xmax = `3D7StartAdjust` + col.4,
# ymin = 1.5, ymax = 2.0),
# data = HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
# geom_rect(aes(xmin = `3D7StartAdjust`, xmax = `3D7StartAdjust` + col.4,
# ymin = -1.0, ymax = -0.5),
# data = HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
# geom_rect(aes(xmin = `HB3StartAdjust`, xmax = `HB3StartAdjust` + col.4,
# ymin = 0.5, ymax = 0.75),
# data = HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
# geom_rect(aes(xmin = `HB3StartAdjust`, xmax = `HB3StartAdjust` + col.4,
# ymin = 0.25, ymax = 0.5),
# data = HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
geom_polygon(data = HB3_chr13_vs_3D7_chr11_nucmer_filt_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
geom_polygon(data = HB3_chr13_vs_3D7_chr13_nucmer_filt_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
sofonias_theme +
scale_y_continuous(breaks = c(-0.75, 0.5, 1.75),
labels = c("3D7-Chr13", "HB3-Chr13", "3D7-Chr11")) +
labs(y = "", x = "") +
theme(panel.border = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 25))
```
```{r}
#| fig-column: screen-inset
print(HB3_chr13_chrom_plot)
```
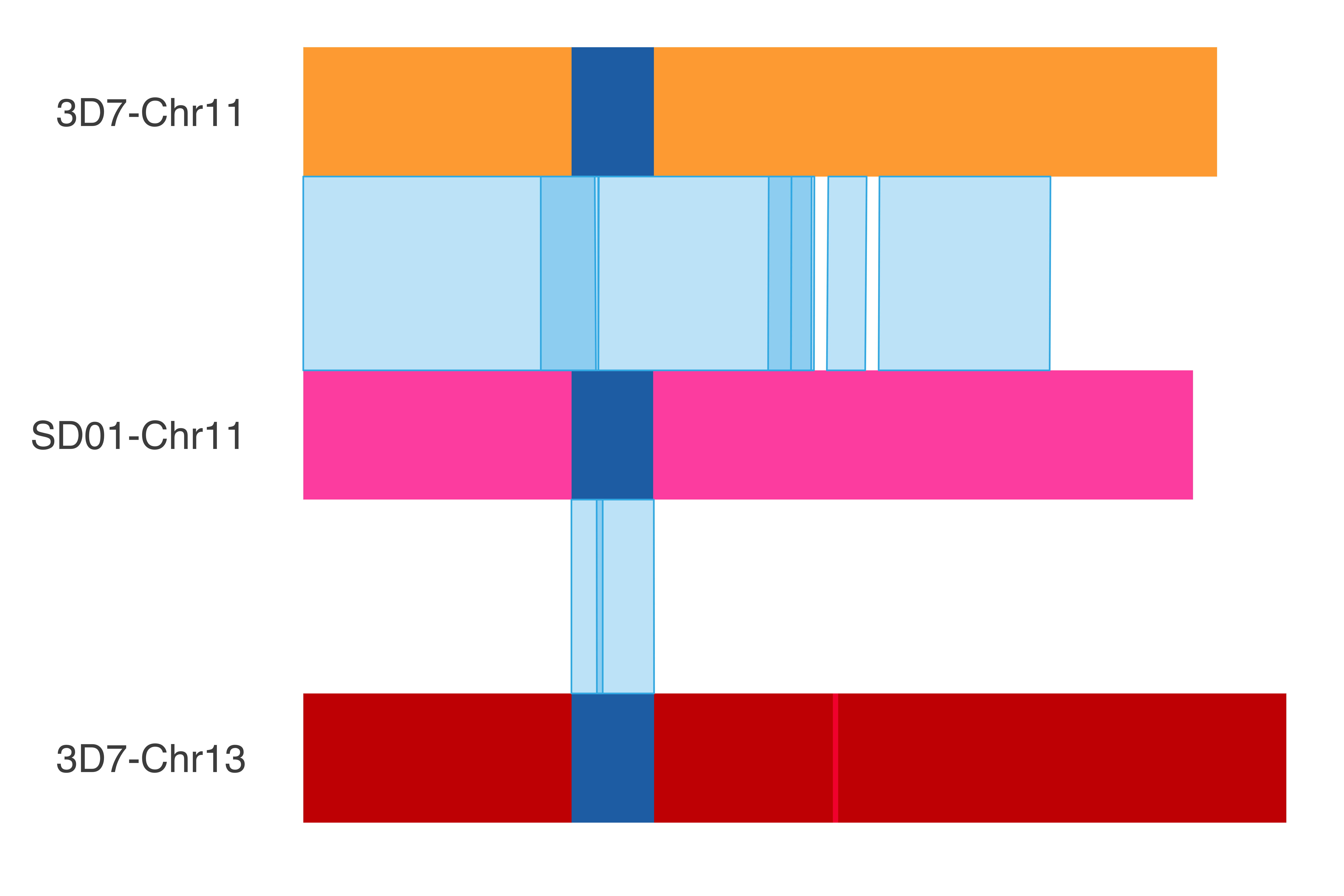
### SD01_chr11
```{r}
SD01_chr11_vs_3D7_chr11_chr13_nucmer = SD01_vs_3D7_nucmer %>%
filter(col.0 %in% c("Pf3D7_11_v3", "Pf3D7_13_v3"),
col.3 %in% c("contig_74_pilon")) %>%
left_join(sharedRegion_SD01 %>%
select(X1, X2) %>%
filter(X1 == "contig_74_pilon") %>%
dplyr::rename(SD01SharedStart = X2,
col.3 = X1)) %>%
left_join(sharedRegion_3D7 %>%
select(chrom, start) %>%
rename(col.0 = chrom,
`3D7SharedStart` = start))
SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt = SD01_chr11_vs_3D7_chr11_chr13_nucmer %>%
filter(col.2 >= (`3D7SharedStart` - backtrace))%>%
filter(actualEnd >= (SD01SharedStart - backtrace)) %>%
mutate(`3D7StartAdjust` = col.1 - (`3D7SharedStart` - backtrace),
SD01StartAdjust = actualStart - (SD01SharedStart - backtrace)) %>%
mutate(`3D7EndAdjust` = `3D7StartAdjust` + col.4,
SD01EndAdjust = SD01StartAdjust + col.4) %>%
group_by(col.0, col.1, col.2, col.3, actualStart, actualEnd) %>%
mutate(`3D7StartAdjust` = max(0, `3D7StartAdjust`),
SD01StartAdjust = max(SD01StartAdjust, 0)) %>%
ungroup()
SD01_chr11_chrom = ChromsSD01 %>%
filter(chrom == "contig_74_pilon") %>%
left_join(SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.3, SD01SharedStart) %>%
rename(chrom = col.3) %>%
unique()) %>%
mutate(start = 0, end = length - (SD01SharedStart - backtrace))
sharedRegion_SD01_chr11_chrom = sharedRegion_SD01 %>%
rename(chrom = X1) %>%
filter(chrom == "contig_74_pilon") %>%
left_join(SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.3, SD01SharedStart) %>%
rename(chrom = col.3) %>%
unique()) %>%
mutate(start = X2 - (SD01SharedStart - backtrace), end = X3 - (SD01SharedStart - backtrace))
Chroms3D7_chr11 = Chroms3D7 %>%
filter(chrom == "Pf3D7_11_v3") %>%
left_join(SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = 0, end = length - (`3D7SharedStart` - backtrace))
sharedRegion_3D7_chr11_chrom = sharedRegion_3D7 %>%
rename(X2 = start,
X3 = end) %>%
filter(chrom == "Pf3D7_11_v3") %>%
left_join(SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace), end = X3 - (`3D7SharedStart` - backtrace))
Chroms3D7_chr13 = Chroms3D7 %>%
filter(chrom == "Pf3D7_13_v3")%>%
left_join(SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = 0, end = length - (`3D7SharedStart` - backtrace))
sharedRegion_3D7_chr13_chrom = sharedRegion_3D7 %>%
rename(X2 = start,
X3 = end) %>%
filter(chrom == "Pf3D7_13_v3") %>%
left_join(SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace), end = X3 - (`3D7SharedStart` - backtrace))
hrpIII_3D7_chr13_chrom = hrpIII_3D7 %>%
rename(chrom = X1) %>%
filter(chrom == "Pf3D7_13_v3") %>%
left_join(SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace), end = X3 - (`3D7SharedStart` - backtrace))
SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt_merged = SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt[,]
SD01_chr11_vs_3D7_chr11_nucmer_filt_ribbon = SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(SD01StartAdjust, SD01EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.75) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 1.5)%>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup())
SD01_chr11_vs_3D7_chr13_nucmer_filt_ribbon = SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(SD01StartAdjust, SD01EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.25) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = -0.5)%>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup())
SD01_chr11_chrom_plot = ggplot() +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#FFAA40",
data = Chroms3D7_chr11) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#2370B2",
data = sharedRegion_3D7_chr11_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#CC0200",
data = Chroms3D7_chr13) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1, ymax = -0.5),
fill = "#2370B2",
data = sharedRegion_3D7_chr13_chrom)+
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#F60239",
data = hrpIII_3D7_chr13_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#FF5AAF",
data = SD01_chr11_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#2370B2",
data = sharedRegion_SD01_chr11_chrom) +
# geom_rect(aes(xmin = `3D7StartAdjust`, xmax = `3D7StartAdjust` + col.4,
# ymin = 1.5, ymax = 2.0),
# data = SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
# geom_rect(aes(xmin = `3D7StartAdjust`, xmax = `3D7StartAdjust` + col.4,
# ymin = -1.0, ymax = -0.5),
# data = SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
# geom_rect(aes(xmin = `SD01StartAdjust`, xmax = `SD01StartAdjust` + col.4,
# ymin = 0.5, ymax = 0.75),
# data = SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
# geom_rect(aes(xmin = `SD01StartAdjust`, xmax = `SD01StartAdjust` + col.4,
# ymin = 0.25, ymax = 0.5),
# data = SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
geom_polygon(data = SD01_chr11_vs_3D7_chr11_nucmer_filt_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
geom_polygon(data = SD01_chr11_vs_3D7_chr13_nucmer_filt_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
sofonias_theme +
scale_y_continuous(breaks = c(-0.75, 0.5, 1.75),
labels = c("3D7-Chr13", "SD01-Chr11", "3D7-Chr11")) +
labs(y = "", x = "") +
theme(panel.border = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 25))
```
```{r}
#| fig-column: screen-inset
print(SD01_chr11_chrom_plot)
```
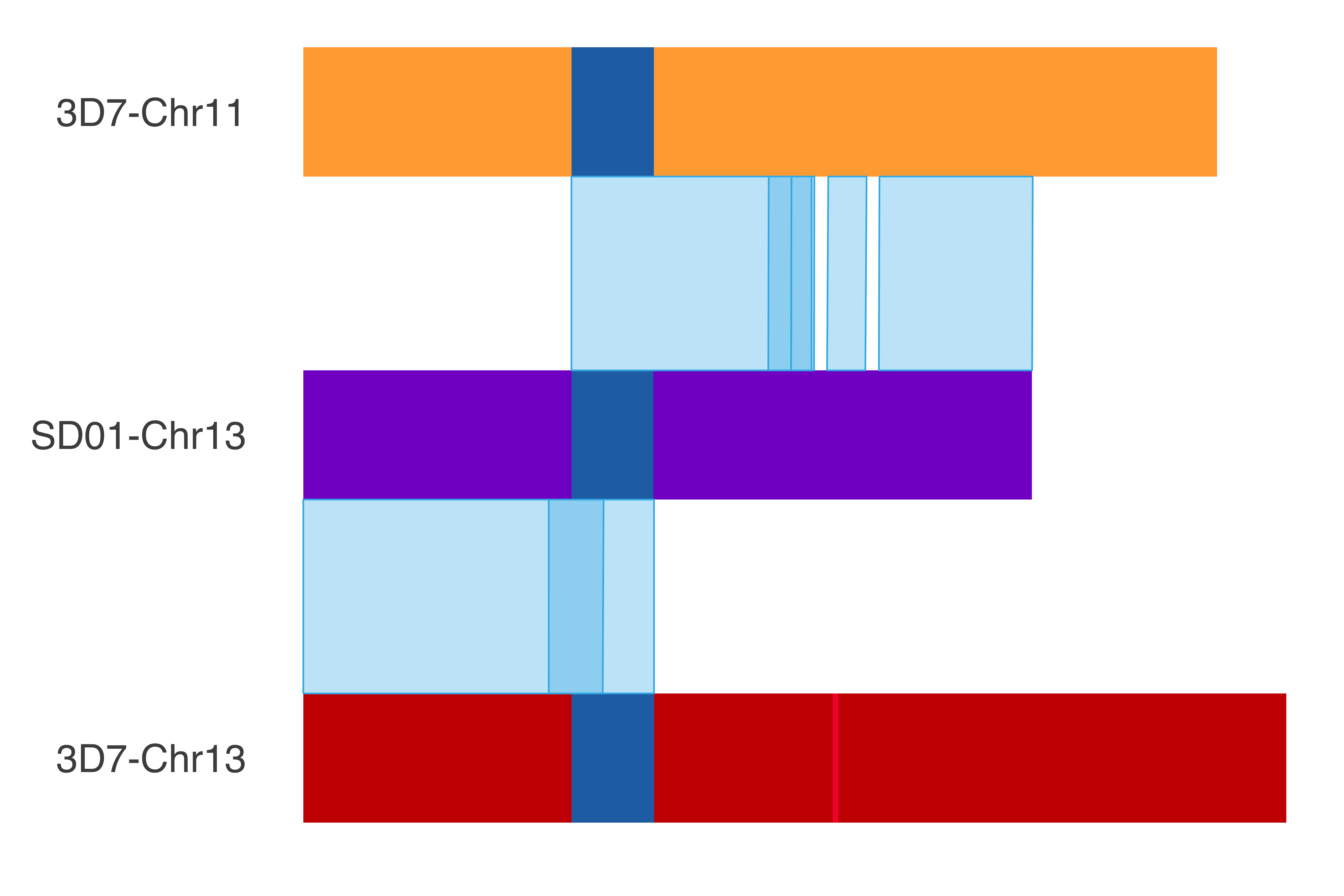
### SD01_chr13
```{r}
SD01_chr13_vs_3D7_chr11_chr13_nucmer = SD01_vs_3D7_nucmer %>%
filter(col.0 %in% c("Pf3D7_11_v3", "Pf3D7_13_v3"),
col.3 %in% c("contig_73_chr13_pilon")) %>%
left_join(sharedRegion_SD01 %>%
select(X1, X2) %>%
filter(X1 == "contig_73_chr13_pilon") %>%
dplyr::rename(SD01SharedStart = X2,
col.3 = X1)) %>%
left_join(sharedRegion_3D7 %>%
select(chrom, start) %>%
rename(col.0 = chrom,
`3D7SharedStart` = start))
SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt = SD01_chr13_vs_3D7_chr11_chr13_nucmer %>%
filter(col.2 >= (`3D7SharedStart` - backtrace))%>%
filter(actualEnd >= (SD01SharedStart - backtrace)) %>%
mutate(`3D7StartAdjust` = col.1 - (`3D7SharedStart` - backtrace),
SD01StartAdjust = actualStart - (SD01SharedStart - backtrace)) %>%
mutate(`3D7EndAdjust` = `3D7StartAdjust` + col.4,
SD01EndAdjust = SD01StartAdjust + col.4) %>%
group_by(col.0, col.1, col.2, col.3, actualStart, actualEnd) %>%
mutate(`3D7StartAdjust` = max(0, `3D7StartAdjust`),
SD01StartAdjust = max(SD01StartAdjust, 0)) %>%
ungroup()
SD01_chr13_chrom = ChromsSD01 %>%
filter(chrom == "contig_73_chr13_pilon") %>%
left_join(SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.3, SD01SharedStart) %>%
rename(chrom = col.3) %>%
unique()) %>%
mutate(start = 0, end = length - (SD01SharedStart - backtrace))
sharedRegion_SD01_chr13_chrom = sharedRegion_SD01 %>%
rename(chrom = X1) %>%
filter(chrom == "contig_73_chr13_pilon") %>%
left_join(SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.3, SD01SharedStart) %>%
rename(chrom = col.3) %>%
unique()) %>%
mutate(start = X2 - (SD01SharedStart - backtrace), end = X3 - (SD01SharedStart - backtrace))
Chroms3D7_chr11 = Chroms3D7 %>%
filter(chrom == "Pf3D7_11_v3") %>%
left_join(SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = 0, end = length - (`3D7SharedStart` - backtrace))
sharedRegion_3D7_chr11_chrom = sharedRegion_3D7 %>%
rename(X2 = start,
X3 = end) %>%
filter(chrom == "Pf3D7_11_v3") %>%
left_join(SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace), end = X3 - (`3D7SharedStart` - backtrace))
Chroms3D7_chr13 = Chroms3D7 %>%
filter(chrom == "Pf3D7_13_v3")%>%
left_join(SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = 0, end = length - (`3D7SharedStart` - backtrace))
sharedRegion_3D7_chr13_chrom = sharedRegion_3D7 %>%
rename(X2 = start,
X3 = end) %>%
filter(chrom == "Pf3D7_13_v3") %>%
left_join(SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace), end = X3 - (`3D7SharedStart` - backtrace))
hrpIII_3D7_chr13_chrom = hrpIII_3D7 %>%
rename(chrom = X1) %>%
filter(chrom == "Pf3D7_13_v3") %>%
left_join(SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
select(col.0, `3D7SharedStart`) %>%
rename(chrom = col.0) %>%
unique()) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace), end = X3 - (`3D7SharedStart` - backtrace))
SD01_chr13_vs_3D7_chr11_nucmer_filt_ribbon = SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(SD01StartAdjust, SD01EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.75) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 1.5)%>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup())
SD01_chr13_vs_3D7_chr13_nucmer_filt_ribbon = SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(SD01StartAdjust, SD01EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.25) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)%>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = -0.5)%>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup())
SD01_chr13_chrom_plot = ggplot() +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#FFAA40",
data = Chroms3D7_chr11) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#2370B2",
data = sharedRegion_3D7_chr11_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#CC0200",
data = Chroms3D7_chr13) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1, ymax = -0.5),
fill = "#2370B2",
data = sharedRegion_3D7_chr13_chrom)+
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#F60239",
data = hrpIII_3D7_chr13_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#8400CD",
data = SD01_chr13_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#2370B2",
data = sharedRegion_SD01_chr13_chrom) +
# geom_rect(aes(xmin = `3D7StartAdjust`, xmax = `3D7StartAdjust` + col.4,
# ymin = 1.5, ymax = 2.0),
# data = SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
# geom_rect(aes(xmin = `3D7StartAdjust`, xmax = `3D7StartAdjust` + col.4,
# ymin = -1.0, ymax = -0.5),
# data = SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
# geom_rect(aes(xmin = `SD01StartAdjust`, xmax = `SD01StartAdjust` + col.4,
# ymin = 0.5, ymax = 0.75),
# data = SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
# geom_rect(aes(xmin = `SD01StartAdjust`, xmax = `SD01StartAdjust` + col.4,
# ymin = 0.25, ymax = 0.5),
# data = SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
# filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff)) +
geom_polygon(data = SD01_chr13_vs_3D7_chr11_nucmer_filt_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
geom_polygon(data = SD01_chr13_vs_3D7_chr13_nucmer_filt_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
sofonias_theme +
scale_y_continuous(breaks = c(-0.75, 0.5, 1.75),
labels = c("3D7-Chr13", "SD01-Chr13", "3D7-Chr11")) +
labs(y = "", x = "") +
theme(panel.border = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 25))
```
```{r}
#| fig-column: screen-inset
print(SD01_chr13_chrom_plot)
```
```{r, results='hide'}
pdf("HB3_chr11_chrom_plot_vs_3D7_chr11_chr13.pdf", width = 8, height = 3, useDingbats = F)
print(HB3_chr11_chrom_plot)
dev.off()
pdf("HB3_chr13_chrom_plot_vs_3D7_chr11_chr13.pdf", width = 8, height = 3, useDingbats = F)
print(HB3_chr13_chrom_plot)
dev.off()
pdf("SD01_chr11_chrom_plot_vs_3D7_chr11_chr13.pdf", width = 8, height = 3, useDingbats = F)
print(SD01_chr11_chrom_plot)
dev.off()
pdf("SD01_chr13_chrom_plot_vs_3D7_chr11_chr13.pdf", width = 8, height = 3, useDingbats = F)
print(SD01_chr13_chrom_plot)
dev.off()
```
```{r}
library(cowplot)
## HB3_chr11_vs_3D7_chr11
HB3_chr11_vs_3D7_chr11_nucmer_filt_select = HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff) %>%
select(HB3StartAdjust, HB3EndAdjust, `3D7StartAdjust`, `3D7EndAdjust`, perID) %>%
arrange(`3D7StartAdjust`, `3D7EndAdjust`)
HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged = HB3_chr11_vs_3D7_chr11_nucmer_filt_select[1,]
for(row in 2:nrow(HB3_chr11_vs_3D7_chr11_nucmer_filt_select)){
if(HB3_chr11_vs_3D7_chr11_nucmer_filt_select$`3D7StartAdjust`[row] < HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged)]){
HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged)] = HB3_chr11_vs_3D7_chr11_nucmer_filt_select$`3D7EndAdjust`[row]
HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged$`HB3EndAdjust`[nrow(HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged)] = HB3_chr11_vs_3D7_chr11_nucmer_filt_select$`HB3EndAdjust`[row]
}else{
HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged = bind_rows(
HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged,
HB3_chr11_vs_3D7_chr11_nucmer_filt_select[row,]
)
}
}
HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged_ribbon = HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(HB3StartAdjust, HB3EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.75) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(
HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 1.5) %>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup()
)
## HB3_chr11_vs_3D7_chr13
HB3_chr11_vs_3D7_chr13_nucmer_filt_select = HB3_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff) %>%
select(HB3StartAdjust, HB3EndAdjust, `3D7StartAdjust`, `3D7EndAdjust`, perID) %>%
arrange(`3D7StartAdjust`, `3D7EndAdjust`)
HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged = HB3_chr11_vs_3D7_chr13_nucmer_filt_select[1,]
if(nrow(HB3_chr11_vs_3D7_chr13_nucmer_filt_select) > 1){
for(row in 2:nrow(HB3_chr11_vs_3D7_chr13_nucmer_filt_select)){
if(HB3_chr11_vs_3D7_chr13_nucmer_filt_select$`3D7StartAdjust`[row] < HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged)]){
HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged)] = HB3_chr11_vs_3D7_chr13_nucmer_filt_select$`3D7EndAdjust`[row]
HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged$`HB3EndAdjust`[nrow(HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged)] = HB3_chr11_vs_3D7_chr13_nucmer_filt_select$`HB3EndAdjust`[row]
}else{
HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged = bind_rows(
HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged,
HB3_chr11_vs_3D7_chr13_nucmer_filt_select[row,]
)
}
}
}
HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged_ribbon = HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(HB3StartAdjust, HB3EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.25) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(
HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = -0.5)%>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup()
)
## HB3_chr13_vs_3D7_chr11
HB3_chr13_vs_3D7_chr11_nucmer_filt_select = HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff) %>%
select(HB3StartAdjust, HB3EndAdjust, `3D7StartAdjust`, `3D7EndAdjust`, perID) %>%
arrange(`3D7StartAdjust`, `3D7EndAdjust`)
HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged = HB3_chr13_vs_3D7_chr11_nucmer_filt_select[1,]
if(nrow(HB3_chr13_vs_3D7_chr11_nucmer_filt_select) > 1){
for(row in 2:nrow(HB3_chr13_vs_3D7_chr11_nucmer_filt_select)){
if(HB3_chr13_vs_3D7_chr11_nucmer_filt_select$`3D7StartAdjust`[row] < HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged)]){
HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged)] = HB3_chr13_vs_3D7_chr11_nucmer_filt_select$`3D7EndAdjust`[row]
HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged$`HB3EndAdjust`[nrow(HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged)] = HB3_chr13_vs_3D7_chr11_nucmer_filt_select$`HB3EndAdjust`[row]
}else{
HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged = bind_rows(
HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged,
HB3_chr13_vs_3D7_chr11_nucmer_filt_select[row,]
)
}
}
}
HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged_ribbon = HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(HB3StartAdjust, HB3EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.75) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(
HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 1.5) %>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup()
)
## HB3_chr13_vs_3D7_chr13
HB3_chr13_vs_3D7_chr13_nucmer_filt_select = HB3_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff) %>%
select(HB3StartAdjust, HB3EndAdjust, `3D7StartAdjust`, `3D7EndAdjust`, perID) %>%
arrange(`3D7StartAdjust`, `3D7EndAdjust`)
HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged = HB3_chr13_vs_3D7_chr13_nucmer_filt_select[1,]
if(nrow(HB3_chr13_vs_3D7_chr13_nucmer_filt_select) > 1){
for(row in 2:nrow(HB3_chr13_vs_3D7_chr13_nucmer_filt_select)){
if(HB3_chr13_vs_3D7_chr13_nucmer_filt_select$`3D7StartAdjust`[row] < HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged)]){
HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged)] = HB3_chr13_vs_3D7_chr13_nucmer_filt_select$`3D7EndAdjust`[row]
HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged$`HB3EndAdjust`[nrow(HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged)] = HB3_chr13_vs_3D7_chr13_nucmer_filt_select$`HB3EndAdjust`[row]
}else{
HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged = bind_rows(
HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged,
HB3_chr13_vs_3D7_chr13_nucmer_filt_select[row,]
)
}
}
}
HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged_ribbon = HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(HB3StartAdjust, HB3EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.25) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(
HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = -0.5)%>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup()
)
## SD01_chr11_vs_3D7_chr11
SD01_chr11_vs_3D7_chr11_nucmer_filt_select = SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff) %>%
select(SD01StartAdjust, SD01EndAdjust, `3D7StartAdjust`, `3D7EndAdjust`, perID) %>%
arrange(`3D7StartAdjust`, `3D7EndAdjust`)
SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged = SD01_chr11_vs_3D7_chr11_nucmer_filt_select[1,]
for(row in 2:nrow(SD01_chr11_vs_3D7_chr11_nucmer_filt_select)){
if(SD01_chr11_vs_3D7_chr11_nucmer_filt_select$`3D7StartAdjust`[row] < SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged)]){
SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged)] = SD01_chr11_vs_3D7_chr11_nucmer_filt_select$`3D7EndAdjust`[row]
SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged$`SD01EndAdjust`[nrow(SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged)] = SD01_chr11_vs_3D7_chr11_nucmer_filt_select$`SD01EndAdjust`[row]
}else{
SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged = bind_rows(
SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged,
SD01_chr11_vs_3D7_chr11_nucmer_filt_select[row,]
)
}
}
SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged_ribbon = SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(SD01StartAdjust, SD01EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.75) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(
SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 1.5) %>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup()
)
## SD01_chr11_vs_3D7_chr13
SD01_chr11_vs_3D7_chr13_nucmer_filt_select = SD01_chr11_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff) %>%
select(SD01StartAdjust, SD01EndAdjust, `3D7StartAdjust`, `3D7EndAdjust`, perID) %>%
arrange(`3D7StartAdjust`, `3D7EndAdjust`)
SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged = SD01_chr11_vs_3D7_chr13_nucmer_filt_select[1,]
if(nrow(SD01_chr11_vs_3D7_chr13_nucmer_filt_select) > 1){
for(row in 2:nrow(SD01_chr11_vs_3D7_chr13_nucmer_filt_select)){
if(SD01_chr11_vs_3D7_chr13_nucmer_filt_select$`3D7StartAdjust`[row] < SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged)]){
SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged)] = SD01_chr11_vs_3D7_chr13_nucmer_filt_select$`3D7EndAdjust`[row]
SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged$`SD01EndAdjust`[nrow(SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged)] = SD01_chr11_vs_3D7_chr13_nucmer_filt_select$`SD01EndAdjust`[row]
}else{
SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged = bind_rows(
SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged,
SD01_chr11_vs_3D7_chr13_nucmer_filt_select[row,]
)
}
}
}
SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged_ribbon = SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(SD01StartAdjust, SD01EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.25) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(
SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = -0.5)%>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup()
)
## SD01_chr13_vs_3D7_chr11
SD01_chr13_vs_3D7_chr11_nucmer_filt_select = SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_11_v3", perID > perIDCutOff, col.4 > lengthCutOff) %>%
select(SD01StartAdjust, SD01EndAdjust, `3D7StartAdjust`, `3D7EndAdjust`, perID) %>%
arrange(`3D7StartAdjust`, `3D7EndAdjust`)
SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged = SD01_chr13_vs_3D7_chr11_nucmer_filt_select[1,]
if(nrow(SD01_chr13_vs_3D7_chr11_nucmer_filt_select) > 1){
for(row in 2:nrow(SD01_chr13_vs_3D7_chr11_nucmer_filt_select)){
if(SD01_chr13_vs_3D7_chr11_nucmer_filt_select$`3D7StartAdjust`[row] < SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged)]){
SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged)] = SD01_chr13_vs_3D7_chr11_nucmer_filt_select$`3D7EndAdjust`[row]
SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged$`SD01EndAdjust`[nrow(SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged)] = SD01_chr13_vs_3D7_chr11_nucmer_filt_select$`SD01EndAdjust`[row]
}else{
SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged = bind_rows(
SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged,
SD01_chr13_vs_3D7_chr11_nucmer_filt_select[row,]
)
}
}
}
SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged_ribbon = SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(SD01StartAdjust, SD01EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.75) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(
SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 1.5) %>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup()
)
## SD01_chr13_vs_3D7_chr13
SD01_chr13_vs_3D7_chr13_nucmer_filt_select = SD01_chr13_vs_3D7_chr11_chr13_nucmer_filt %>%
filter(col.0 == "Pf3D7_13_v3", perID > perIDCutOff, col.4 > lengthCutOff) %>%
select(SD01StartAdjust, SD01EndAdjust, `3D7StartAdjust`, `3D7EndAdjust`, perID) %>%
arrange(`3D7StartAdjust`, `3D7EndAdjust`)
SD01_chr13_vs_3D7_chr13_nucmer_filt_select_merged = SD01_chr13_vs_3D7_chr13_nucmer_filt_select[1,]
if(nrow(SD01_chr13_vs_3D7_chr13_nucmer_filt_select) > 1){
for(row in 2:nrow(SD01_chr13_vs_3D7_chr13_nucmer_filt_select)){
if(SD01_chr13_vs_3D7_chr13_nucmer_filt_select$`3D7StartAdjust`[row] < SD01_chr13_vs_3D7_chr13_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(SD01_chr13_vs_3D7_chr13_nucmer_filt_select_merged)]){
SD01_chr13_vs_3D7_chr13_nucmer_filt_select_merged$`3D7EndAdjust`[nrow(SD01_chr13_vs_3D7_chr13_nucmer_filt_select_merged)] = SD01_chr13_vs_3D7_chr13_nucmer_filt_select$`3D7EndAdjust`[row]
SD01_chr13_vs_3D7_chr13_nucmer_filt_select_merged$`SD01EndAdjust`[nrow(SD01_chr13_vs_3D7_chr13_nucmer_filt_select_merged)] = SD01_chr13_vs_3D7_chr13_nucmer_filt_select$`SD01EndAdjust`[row]
}else{
SD01_chr13_vs_3D7_chr13_nucmer_filt_select_merged = bind_rows(
SD01_chr13_vs_3D7_chr13_nucmer_filt_select_merged,
SD01_chr13_vs_3D7_chr13_nucmer_filt_select[row,]
)
}
}
}
SD01_chr13_vs_3D7_chr13_nucmer_filt_select_merged_ribbon = SD01_chr13_vs_3D7_chr13_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(SD01StartAdjust, SD01EndAdjust, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = 0.25) %>%
arrange(rowid, x) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number())) %>%
ungroup() %>%
bind_rows(
HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged %>%
mutate(rowid = row_number()) %>%
select(`3D7StartAdjust`, `3D7EndAdjust`, perID, rowid) %>%
gather(key, x, 1:2) %>%
mutate(y = -0.5)%>%
arrange(rowid, desc(x)) %>%
group_by(rowid) %>%
mutate(point = paste0("point", row_number() + 2)) %>%
ungroup()
)
```
## Combining into one plot
```{r}
threed7_tares_chr11 = threed7_tares %>%
filter(X1 == "Pf3D7_11_v3") %>%
rename(chrom = X1) %>%
left_join(Chroms3D7_chr11 %>%
select(chrom, `3D7SharedStart`)) %>%
filter(X2 > `3D7SharedStart`) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace),
end = X3 - (`3D7SharedStart` - backtrace))
threed7_tares_chr13 = threed7_tares %>%
filter(X1 == "Pf3D7_13_v3") %>%
rename(chrom = X1) %>%
left_join(Chroms3D7_chr13 %>%
select(chrom, `3D7SharedStart`)) %>%
filter(X2 > `3D7SharedStart`) %>%
mutate(start = X2 - (`3D7SharedStart` - backtrace),
end = X3 - (`3D7SharedStart` - backtrace))
hb3_tares_chr11 = hb3_tares %>%
filter(X1 == "PfHB3nano_11") %>%
rename(chrom = X1) %>%
left_join(HB3_chr11_chrom %>%
select(chrom, `HB3SharedStart`)) %>%
filter(X2 > `HB3SharedStart`) %>%
mutate(start = X2 - (`HB3SharedStart` - backtrace),
end = X3 - (`HB3SharedStart` - backtrace))
hb3_tares_chr13 = hb3_tares %>%
filter(X1 == "PfHB3nano_13") %>%
rename(chrom = X1) %>%
left_join(HB3_chr13_chrom %>%
select(chrom, `HB3SharedStart`)) %>%
filter(X2 > `HB3SharedStart`) %>%
mutate(start = X2 - (`HB3SharedStart` - backtrace),
end = X3 - (`HB3SharedStart` - backtrace))
allChroms = bind_rows(
Chroms3D7_chr11,
Chroms3D7_chr13,
HB3_chr11_chrom,
HB3_chr13_chrom,
SD01_chr11_chrom,
SD01_chr13_chrom
)
allChroms_empty = allChroms %>%
select(start, end) %>%
summarise(end = max(end))
forDots = allChroms_empty$end/100
HB3_chr11_chrom_plot = ggplot() +
geom_point(
aes(x = x, y = y),
data = tibble(
x = rep(c(-forDots -forDots * 0.75, -forDots* 0.75 -10 * forDots, -forDots* 0.75 -20 * forDots), 3),
y = c(
-0.75, -0.75, -0.75,
0.5, 0.5, 0.5,
1.75, 1.75, 1.75
)
),
size = 2
) +
geom_rect(aes(xmin = 0, xmax= end,
ymin = 0, ymax = 1),
fill = "#00000000",
data = allChroms_empty) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#CCCCCC",
data = Chroms3D7_chr11) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#2370B2",
data = sharedRegion_3D7_chr11_chrom) +
geom_rect(aes(
xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0,
),
fill = "#E20134",
data = threed7_tares_chr11) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#555555",
data = Chroms3D7_chr13) +
geom_rect(aes(
xmin = start, xmax = end,
ymin = -1, ymax = -0.5,
),
fill = "#E20134",
data = threed7_tares_chr13) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1, ymax = -0.5),
fill = "#2370B2",
data = sharedRegion_3D7_chr13_chrom)+
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#4CD96D",
data = hrpIII_3D7_chr13_chrom) +
geom_text(aes(x = start + (end - start)/2, y = -1.25), label = "pfhrp3",
data = hrpIII_3D7_chr13_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#999999",
data = HB3_chr11_chrom) +
geom_rect(aes(
xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75,
),
fill = "#E20134",
data = hb3_tares_chr11) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#2370B2",
data = sharedRegion_HB3_chr11_chrom) +
geom_polygon(data = HB3_chr11_vs_3D7_chr11_nucmer_filt_select_merged_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
geom_polygon(data = HB3_chr11_vs_3D7_chr13_nucmer_filt_select_merged_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
sofonias_theme +
scale_y_continuous(breaks = c(-0.75, 0.5, 1.75),
labels = c("3D7-Chr13", "HB3-Chr11", "3D7-Chr11"),
limits = c(-1.5, 2),
expand = c(0,0)) +
labs(y = "", x = "") +
theme(panel.border = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 25))
HB3_chr13_chrom_plot = ggplot() +
geom_point(
aes(x = x, y = y),
data = tibble(
x = rep(c(-forDots -forDots * 0.75, -forDots* 0.75 -10 * forDots, -forDots* 0.75 -20 * forDots), 3),
y = c(
-0.75, -0.75, -0.75,
0.5, 0.5, 0.5,
1.75, 1.75, 1.75
)
),
size = 2
) +
geom_rect(aes(xmin = 0, xmax= end,
ymin = 0, ymax = 1),
fill = "#00000000",
data = allChroms_empty)+
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#CCCCCC",
data = Chroms3D7_chr11) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#2370B2",
data = sharedRegion_3D7_chr11_chrom) +
geom_rect(aes(
xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0,
),
fill = "#E20134",
data = threed7_tares_chr11) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#555555",
data = Chroms3D7_chr13) +
geom_rect(aes(
xmin = start, xmax = end,
ymin = -1, ymax = -0.5,
),
fill = "#E20134",
data = threed7_tares_chr13) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1, ymax = -0.5),
fill = "#2370B2",
data = sharedRegion_3D7_chr13_chrom)+
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#4CD96D",
data = hrpIII_3D7_chr13_chrom) +
geom_text(aes(x = start + (end - start)/2, y = -1.25), label = "pfhrp3",
data = hrpIII_3D7_chr13_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#999999",
data = HB3_chr13_chrom) +
geom_rect(aes(
xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75,
),
fill = "#E20134",
data = hb3_tares_chr13)+
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#2370B2",
data = sharedRegion_HB3_chr13_chrom) +
geom_polygon(data = HB3_chr13_vs_3D7_chr11_nucmer_filt_select_merged_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
geom_polygon(data = HB3_chr13_vs_3D7_chr13_nucmer_filt_select_merged_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
sofonias_theme +
scale_y_continuous(breaks = c(-0.75, 0.5, 1.75),
labels = c("3D7-Chr13", "HB3-Chr13", "3D7-Chr11"),
limits = c(-1.5, 2),
expand = c(0,0)) +
labs(y = "", x = "") +
theme(panel.border = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 25))
SD01_chr11_chrom_plot = ggplot() +
geom_point(
aes(x = x, y = y),
data = tibble(
x = rep(c(-forDots -forDots * 0.75, -forDots* 0.75 -10 * forDots, -forDots* 0.75 -20 * forDots), 3),
y = c(
-0.75, -0.75, -0.75,
0.5, 0.5, 0.5,
1.75, 1.75, 1.75
)
),
size = 2
) +
geom_rect(aes(xmin = 0, xmax= end,
ymin = 0, ymax = 1),
fill = "#00000000",
data = allChroms_empty)+
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#CCCCCC",
data = Chroms3D7_chr11) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#2370B2",
data = sharedRegion_3D7_chr11_chrom) +
geom_rect(aes(
xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0,
),
fill = "#E20134",
data = threed7_tares_chr11)+
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#555555",
data = Chroms3D7_chr13) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1, ymax = -0.5),
fill = "#2370B2",
data = sharedRegion_3D7_chr13_chrom) +
geom_rect(aes(
xmin = start, xmax = end,
ymin = -1, ymax = -0.5,
),
fill = "#E20134",
data = threed7_tares_chr13)+
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#4CD96D",
data = hrpIII_3D7_chr13_chrom) +
geom_text(aes(x = start + (end - start)/2, y = -1.25), label = "pfhrp3",
data = hrpIII_3D7_chr13_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#999999",
data = SD01_chr11_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#2370B2",
data = sharedRegion_SD01_chr11_chrom) +
geom_polygon(data = SD01_chr11_vs_3D7_chr11_nucmer_filt_select_merged_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
geom_polygon(data = SD01_chr11_vs_3D7_chr13_nucmer_filt_select_merged_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
sofonias_theme +
scale_y_continuous(breaks = c(-0.75, 0.5, 1.75),
labels = c("3D7-Chr13", "SD01-Chr11", "3D7-Chr11"),
limits = c(-1.5, 2),
expand = c(0,0)) +
labs(y = "", x = "") +
theme(panel.border = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 25))
SD01_chr13_chrom_plot = ggplot() +
geom_point(
aes(x = x, y = y),
data = tibble(
x = rep(c(-forDots -forDots * 0.75, -forDots* 0.75 -10 * forDots, -forDots* 0.75 -20 * forDots), 3),
y = c(
-0.75, -0.75, -0.75,
0.5, 0.5, 0.5,
1.75, 1.75, 1.75
)
),
size = 2
) +
geom_rect(aes(xmin = 0, xmax= end,
ymin = 0, ymax = 1),
fill = "#00000000",
data = allChroms_empty)+
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#CCCCCC",
data = Chroms3D7_chr11) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0),
fill = "#2370B2",
data = sharedRegion_3D7_chr11_chrom) +
geom_rect(aes(
xmin = start, xmax = end,
ymin = 1.5, ymax = 2.0,
),
fill = "#E20134",
data = threed7_tares_chr11)+
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#555555",
data = Chroms3D7_chr13) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1, ymax = -0.5),
fill = "#2370B2",
data = sharedRegion_3D7_chr13_chrom) +
geom_rect(aes(
xmin = start, xmax = end,
ymin = -1, ymax = -0.5,
),
fill = "#E20134",
data = threed7_tares_chr13) +
geom_rect(aes(xmin = start, xmax = end,
ymin = -1.0, ymax = -0.5),
fill = "#4CD96D",
data = hrpIII_3D7_chr13_chrom) +
geom_text(aes(x = start + (end - start)/2, y = -1.25), label = "pfhrp3",
data = hrpIII_3D7_chr13_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#999999",
data = SD01_chr13_chrom) +
geom_rect(aes(xmin = start, xmax = end,
ymin = 0.25, ymax = 0.75),
fill = "#2370B2",
data = sharedRegion_SD01_chr13_chrom) +
geom_polygon(data = SD01_chr13_vs_3D7_chr11_nucmer_filt_select_merged_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
geom_polygon(data = SD01_chr13_vs_3D7_chr13_nucmer_filt_select_merged_ribbon ,
mapping = aes(x = x, y = y, group = rowid), fill = "#3AB7E955", color = "#3AB7E9") +
sofonias_theme +
scale_y_continuous(breaks = c(-0.75, 0.5, 1.75),
labels = c("3D7-Chr13", "SD01-Chr13", "3D7-Chr11"),
limits = c(-1.5, 2),
expand = c(0,0)) +
labs(y = "", x = "") +
theme(panel.border = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 25))
four_by_four_plot = ggdraw() +
draw_plot(HB3_chr11_chrom_plot +
theme(axis.text.y = element_text(size = 10)), 0.0, 0.5, .5, .5) +
draw_plot(HB3_chr13_chrom_plot +
theme(axis.text.y = element_text(size = 10)), 0.0, 0.0, .5, .5) +
draw_plot(SD01_chr11_chrom_plot +
theme(axis.text.y = element_text(size = 10)), 0.5, 0.5, .5, .5) +
draw_plot(SD01_chr13_chrom_plot +
theme(axis.text.y = element_text(size = 10)), 0.5, 0.0, .5, .5)
```
```{r}
#| fig-column: screen-inset
print(four_by_four_plot)
```
```{r}
pdf("SD01_HB3_chrs11_13_against_3D7.pdf", width = 8, height = 3, useDingbats = F)
print(four_by_four_plot)
dev.off()
```
```{r}
#| results: asis
#| echo: false
cat(createDownloadLink("SD01_HB3_chrs11_13_against_3D7.pdf"))
```