Code
for x in /tank/data/genomes/plasmodium/genomes/pf/genomes/*.fasta; do elucidator splitFile --fasta ${x} --overWrite --trimAtWhiteSpace ; done;These strains have complete chromosome 11s and 13s
Pf3D7
Pf7G8
PfCD01
PfDd2
PfGA01
PfGB4
PfGN01
PfKE01
PfKH02
PfSN01
strains = c("Pf3D7","Pf7G8","PfCD01","PfDd2","PfGA01","PfGB4","PfGN01","PfKE01","PfKH02","PfSN01")
cat(strains, sep = "\n", file = "stableStrains.txt")
strainsDf = tibble(strain = strains)
strainsDf = strainsDf %>%
mutate(chrom11 = paste0(strain, "_11*.fasta")) %>%
mutate(chrom13 = paste0(strain, "_13*.fasta"))
mvCmd = paste0("mv ", paste0(strainsDf$chrom11, collapse = " "), " ", paste0(strainsDf$chrom13, collapse = " "), " combinedChrom11_13/")
cat(mvCmd)mv Pf3D7_11*.fasta Pf7G8_11*.fasta PfCD01_11*.fasta PfDd2_11*.fasta PfGA01_11*.fasta PfGB4_11*.fasta PfGN01_11*.fasta PfKE01_11*.fasta PfKH02_11*.fasta PfSN01_11*.fasta Pf3D7_13*.fasta Pf7G8_13*.fasta PfCD01_13*.fasta PfDd2_13*.fasta PfGA01_13*.fasta PfGB4_13*.fasta PfGN01_13*.fasta PfKE01_13*.fasta PfKH02_13*.fasta PfSN01_13*.fasta combinedChrom11_13/elucidator profileSharedKmerBlocks --seq Pf3D7_11_v3.fasta --dout chroms11_13_klen31 --kLen 31 --seqsDir ./ --fasta Pf3D7_11_v3.fasta --overWriteDir
cd chroms11_13_klen31
elucidator bedCoordSort --bed perfect.bed | elucidator extendBedRegions --left 5 --right 5 --bed STDIN | bedtools merge | elucidator bed3ToBed6 --bed STDIN | elucidator filterBedRecordsByLength --bed STDIN --minLen 65 | elucidator bedGetIntersectingGenesInGff --gff /work/nhathawa/genomes/pfGenomes_updated/info//gff/Pf3D7.gff --extraAttributes description --overWrite --bed STDIN --out perfectlyShared_minlen65.bed
elucidator getFastaWithBed --twoBit /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.2bit --overWrite --bed perfectlyShared_minlen65.bed --out perfectlyShared_minlen65
elucidator extractRefSeqsFromGenomes --genomeDir /tank/data/genomes/plasmodium/genomes/pf/genomes/ --gffDir /tank/data/genomes/plasmodium/genomes/pf/info/gff/ --primaryGenome Pf3D7 --bed perfectlyShared_minlen65.bed --outputDir extractedRefSeqs --identity 95 --coverage 95 --numThreads 15perfectlyShared_minlen65_regions = readr::read_tsv("combinedChrom11_13/chroms11_13_klen31/perfectlyShared_minlen65.bed", col_names = F)
perfectlyShared_minlen65_regions = perfectlyShared_minlen65_regions %>%
mutate(X4 = paste0(X4, "-for"))
allRegions = tibble()
for(region in perfectlyShared_minlen65_regions$X4){
for(strain in strains){
fnp= paste0("combinedChrom11_13/chroms11_13_klen31/extractedRefSeqs/", region, "/beds/", strain, "_region.bed")
if(file.exists(fnp)){
currentRegion = readr::read_tsv(fnp, col_names = F, show_col_types = F)
currentRegion$X4 = region
currentRegion$strain = strain
allRegions = bind_rows(
allRegions, currentRegion
)
}
}
}
allRegions_sum = allRegions %>%
group_by(strain, X1) %>%
summarise(start = min(X2),
end = max(X3)) %>%
filter(grepl("_1[13]", X1)) %>%
rename(`#chrom` = X1) %>%
mutate(start = start - 5000) %>%
mutate(end = end + 5000) %>%
mutate(name = paste0(`#chrom`, "-", start, "-", end),
len = end - start,
strand = "+")
for(strainName in strains){
allRegions_sum_byStrain = allRegions_sum %>%
filter(strain == strainName)
allRegions_sum_byStrain = allRegions_sum_byStrain %>%
group_by() %>%
select(-strain)
write_tsv(allRegions_sum_byStrain, paste0("combinedChrom11_13/chroms11_13_regions/", strainName, ".bed"))
}
allRegions_sum_exact = allRegions %>%
group_by(strain, X1) %>%
summarise(start = min(X2),
end = max(X3)) %>%
filter(grepl("_1[13]", X1)) %>%
rename(`#chrom` = X1) %>%
mutate(start = start) %>%
mutate(end = end) %>%
mutate(name = paste0(`#chrom`, "-", start, "-", end),
len = end - start,
strand = "+")
for(strainName in strains){
allRegions_sum_byStrain = allRegions_sum_exact %>%
filter(strain == strainName)
allRegions_sum_byStrain = allRegions_sum_byStrain %>%
group_by() %>%
select(-strain)
write_tsv(allRegions_sum_byStrain, paste0("combinedChrom11_13/chroms11_13_regions_exact/", strainName, ".bed"))
}cd chroms11_13_regions_exact
for x in *.bed; do elucidator getFastaWithBed --bed $x --twoBit /tank/data/genomes/plasmodium/genomes/pf/genomes/${x%%.bed}.2bit --overWrite --out ${x%%.bed}.fasta ; done;
mkdir sequences
cd sequences
for x in ../*.fasta; do elucidator splitFile --fasta $x ; done;
elucidator profileSharedKmerBlocks --seq Pf3D7_11_v3-1913023-1938396.fasta --dout chroms11_13_klen31 --kLen 31 --seqsDir ./ --fasta Pf3D7_11_v3-1913023-1938396.fasta --overWriteDircd chroms11_13_regions
mkdir allSequences
for x in Pf*.bed; do elucidator getFastaWithBed --bed $x --twoBit /tank/data/genomes/plasmodium/genomes/pf/genomes/${x%%.bed}.2bit --overWrite --out ${x%%.bed}.fasta ; done;
cat *.fasta > allSequences/all.fasta
cat Pf*.bed > all.bed
cd allSequences
muscle -in all.fasta -out aln_all.fasta
elucidator trimToLen --fasta aln_all.fasta --length 21945 --overWrite --out trimmed_to_21945_aln_all.fasta
elucidator trimFront --fasta trimmed_to_21945_aln_all.fasta --forwardBases 21845 --overWrite --out trimmedFrom_21845_trimmed_to_21945_aln_all.fasta
elucidator sortReads --fasta trimmedFrom_21845_trimmed_to_21945_aln_all.fasta --removeGaps --overWrite
elucidator determineRegionLastz --individual --fasta sorted_trimmedFrom_21845_trimmed_to_21945_aln_all.fasta --genome /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.2bit -out sorted_trimmedFrom_21845_trimmed_to_21945_aln_all.bed
elucidator trimToLen --fasta aln_all.fasta --length 6051 --overWrite --out trimmed_to_6051_aln_all.fasta
elucidator trimFront --fasta trimmed_to_6051_aln_all.fasta --forwardBases 5951 --overWrite --out trimmedFrom_5951_trimmed_to_6051_aln_all.fasta
elucidator sortReads --fasta trimmedFrom_5951_trimmed_to_6051_aln_all.fasta --removeGaps --overWrite
elucidator determineRegionLastz --individual --fasta sorted_trimmedFrom_5951_trimmed_to_6051_aln_all.fasta --genome /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.2bit --overWrite --out sorted_trimmedFrom_5951_trimmed_to_6051_aln_all.bed
elucidatorlab getMalnConservedness --fasta aln_all.fasta --blockSize 10 --overWrite --out aln_all_conserved.tab.txt
elucidatorlab getAlnPosToRealPosTable --fasta aln_all.fasta --overWrite --out aln_all_position_table.tab.txt Pf3D7_11_v3-1918029-1918124–Pf3D7_13_v3-2792022-2792117

Variation in the middle, does not appear to be chromosome specific (though could have been a difference in assembly)

Pf3D7_11_v3-1933291-1933390–Pf3D7_13_v3-2807298-2807397

The region is ~15,360 kp long, so could be hard to get through even with long reads
Plotting the percent conserved per position across the region including the before and after shared segment, all strains appear to start and end at the same position
conservedPosTab = readr::read_tsv("combinedChrom11_13/chroms11_13_regions/allSequences/aln_all_position_table.tab.txt") %>%
filter("Pf3D7_11_v3-1913023-1938396" == name) %>%
rename(pos = alignPosition)
conservedMulti = readr::read_tsv("combinedChrom11_13/chroms11_13_regions/allSequences/aln_all_conserved.tab.txt")
conservedMulti_max = conservedMulti %>%
group_by(pos) %>%
filter(frac == max(frac)) %>%
filter("----------" != block ) %>%
left_join(conservedPosTab) %>%
filter(!is.na(name)) %>%
filter()
blockSize = 30
conservedMulti_max_rounding = tibble(rawPos = 1:(nrow(conservedMulti_max) - blockSize),
realPosition = numeric(nrow(conservedMulti_max) - blockSize),
rounded = numeric(nrow(conservedMulti_max) - blockSize))
for(row in 1:(nrow(conservedMulti_max) - blockSize)){
round = mean(conservedMulti_max$frac[row:(row+blockSize)])
if(round > 1){
print(row)
print(blockSize)
print(conservedMulti_max$frac[row:(row+blockSize)])
print(round)
stop()
}
minPos = min(conservedMulti_max$realPosition[row:(row+blockSize)])
conservedMulti_max_rounding$realPosition[row] = minPos
conservedMulti_max_rounding$rounded[row] = round
}
conservedMulti_max_rounding_filt = conservedMulti_max_rounding %>%
group_by(realPosition) %>%
mutate(id = row_number()) %>%
filter(id == 1) %>%
group_by()ggplot(conservedMulti_max_rounding_filt) +
geom_bar(aes(x = realPosition, y = rounded), stat = "identity")+
sofonias_theme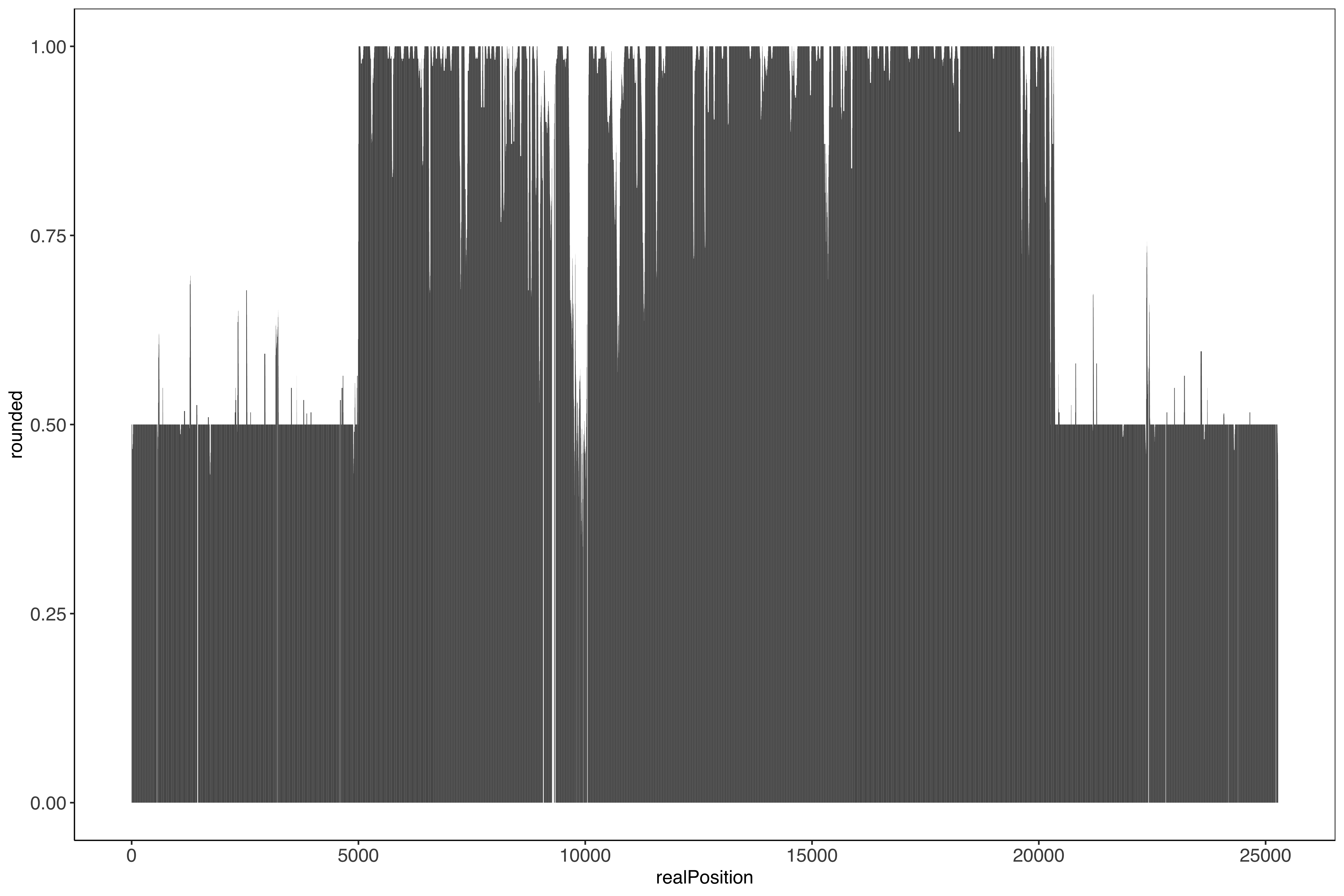
cd chroms11_13_regions_exact
cat *.fasta > all.fasta
elucidator compareAllByAll --fasta all.fasta --verbose --numThreads 12
elucidator createSharedSubSegmentsFromRefSeqs --fasta out.fasta --minimumKlen 12 --dout sharedSubSeqs_occurence6 --filterRegionsWithinRegions --refSeqName Pf3D7_11_v3-1918023-1933396 --overWriteDir --genome /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.fasta --minSubRegionLen 150 --maxSubRegionLen 300 --minBlockRegionLen 30 --doNotCollapseLowFreqNodes --correctionOccurenceCutOff 6 --includeFromSubRegionSize 30
muscle -in all.fasta -out aln_all.fasta
FastTree -nt aln_all.fasta > aln_all.nwk
elucidatorlab getMalnConservedness --fasta aln_all.fasta --blockSize 10 --overWrite --out aln_all_conserved.tab.txt
elucidatorlab getAlnPosToRealPosTable --fasta aln_all.fasta --overWrite --out aln_all_position_table.tab.txt
elucidator multipleAlnProteinToPcaInput --fasta aln_all.fasta --overWrite --out pcaMat_aln_all.fasta
elucidator runTRF --fasta all.fasta --overWriteDir --dout all_trfOutput --supplement --genomicLocation ../all.bed
elucidator bedFilterRegionsCompletelyInOther --bed all_trfOutput/genomic_combined.bed --intersectWithBed all_trfOutput/genomic_combined.bed | elucidator bedAddSmartIDForPlotting --bed STDIN --out all_trfOutput/filtered_genomic_combined.bed --overWrite
elucidator countBasesPerPosition --fasta aln_all.fasta --overWrite --out aln_all_baseCounts.tab.txt
#elucidator createSharedSubSegmentsFromRefSeqs --fasta out.fasta --minimumKlen 12 --dout sharedSubSeqs_occurence6 --filterRegionsWithinRegions --refSeqName Pf3D7_11_v3-1918023-1933396 --overWriteDir --genome /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.fasta --minSubRegionLen 150 --maxSubRegionLen 300 --minBlockRegionLen 30 --doNotCollapseLowFreqNodes --correctionOccurenceCutOff 6 --includeFromSubRegionSize 30
#cd sharedSubSeqs_occurence6/sharedLocs
#elucidator bedGetIntersectingGenesInGff --gff /tank/data/genomes/plasmodium/genomes/pf/info/gff/Pf3D7.gff --extraAttributes description --overWrite --bed 0_ref_sharedLocs_genomic.bed --out 0_ref_sharedLocs_genomic_withGeneInfo.bedThe chromosome 11 and 13 regions appear to be extremely closely related to each other and there is no clear variation specific to chromosome 11 or 13
library(ggtree)
tree = ape::read.tree("combinedChrom11_13/chroms11_13_regions_exact/sequences/aln_all.nwk")
tree$tip.label = gsub("_v3", "", gsub("-.*", "", tree$tip.label))
tipDf = tibble(seq = tree$tip.label) %>%
mutate(chrom = ifelse(grepl("_11", seq), "chrom11", "chrom13"))
tipDf_chrom11 = tipDf %>%
filter(chrom == "chrom11")
tipDf_chrom13 = tipDf %>%
filter(chrom == "chrom13")
groups = list(chrom11 = tipDf_chrom11$seq,
chrom13 = tipDf_chrom13$seq)
tree = groupOTU(tree, groups)
sharedRegion_pacbioAssembliesPlot = ggtree::ggtree(tree, show.legend = FALSE) +
hexpand(.1) +
geom_tiplab(aes(color = group), show.legend = FALSE) +
scale_color_manual(values = c("#005AC8", "#AA0A3C")) print(sharedRegion_pacbioAssembliesPlot)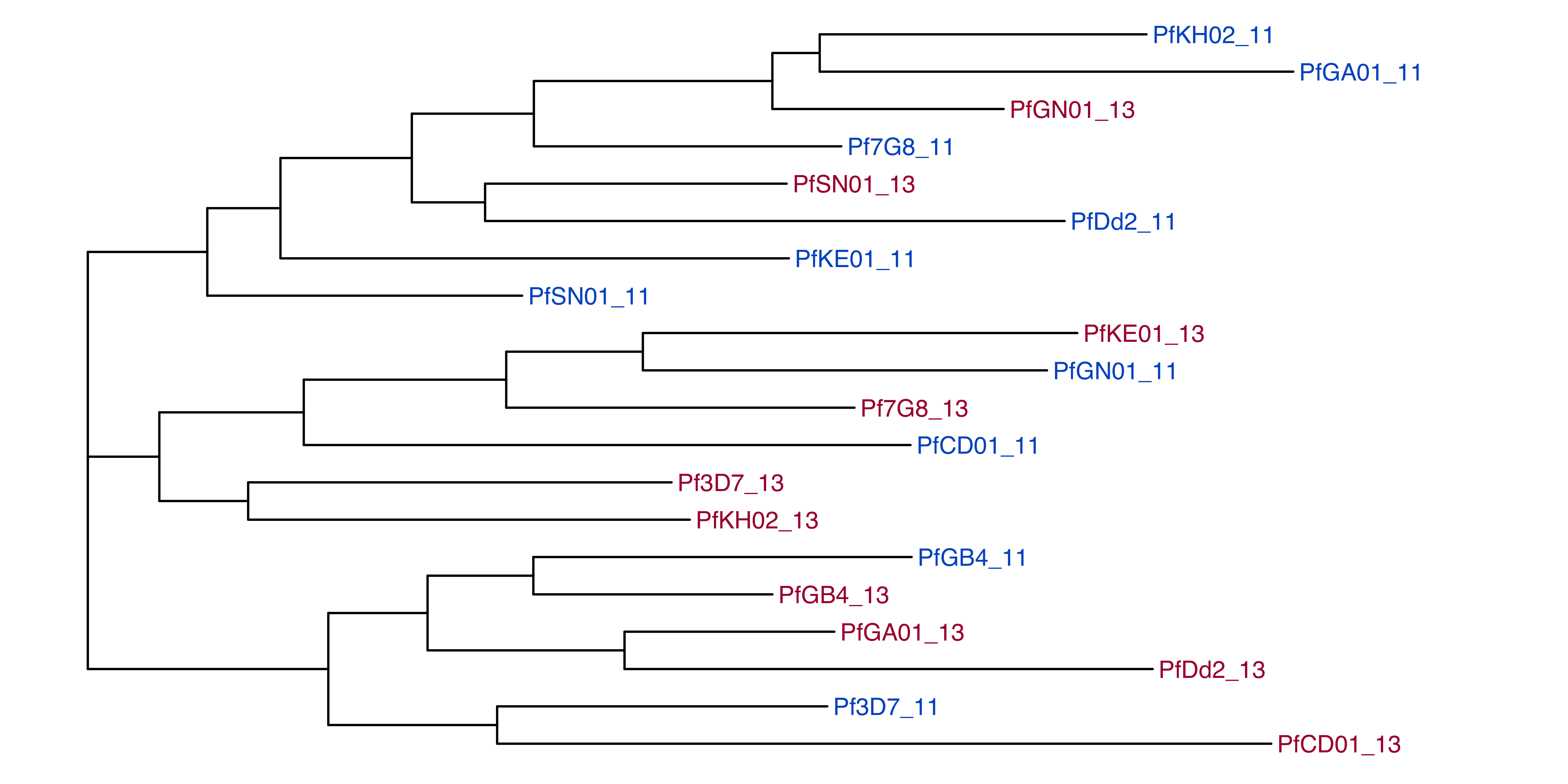
quartz_off_screen
2 All segments between strains are more >99% related to each other
allByAllComp = readr::read_tsv("combinedChrom11_13/chroms11_13_regions_exact/sequences/out.tab.txt")allByAllComp_select = allByAllComp %>%
select(OtherReadId, ReadId, perId) %>%
spread(ReadId, perId)
addRow = matrix(c(colnames(allByAllComp_select)[2], rep(NA, ncol(allByAllComp_select) - 1)), nrow = 1)
colnames(addRow) = colnames(allByAllComp_select)
addRow = as_tibble(addRow)
for(col in 2:ncol(addRow)){
addRow[,col] = as.numeric(addRow[,col])
}
allByAllComp_select = as_tibble(addRow) %>% bind_rows(allByAllComp_select)
allByAllComp_select_mat = as.matrix(allByAllComp_select[,2:ncol(allByAllComp_select)])
rownames(allByAllComp_select_mat) = allByAllComp_select$OtherReadId
allByAllComp_select_mat = as.matrix(as.dist(allByAllComp_select_mat))
allByAllComp_select_mat[0 == allByAllComp_select_mat] = NAlibrary(circlize)
meta = readr::read_tsv("../../meta/metadata/meta.tab.txt") %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
metaByBioSample = readr::read_tsv("../../meta/metadata/metaByBioSample.tab.txt") %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
allByAllComp_select_mat_noLabels = allByAllComp_select_mat
# rownames(allByAllComp_select_mat_noLabels) = NULL
# colnames(allByAllComp_select_mat_noLabels) = NULL
rownames(allByAllComp_select_mat_noLabels) = gsub("Dd2", "DD2", gsub("Pf", "", gsub("_v3", "", gsub("-.*", "", rownames(allByAllComp_select_mat_noLabels)))))
colnames(allByAllComp_select_mat_noLabels) = gsub("Dd2", "DD2", gsub("Pf", "", gsub("_v3", "", gsub("-.*", "", colnames(allByAllComp_select_mat_noLabels)))))
col_fun = colorRamp2(c(
min(allByAllComp_select_mat_noLabels, na.rm = T),
min(allByAllComp_select_mat_noLabels, na.rm = T) + (
max(allByAllComp_select_mat_noLabels, na.rm = T) - min(allByAllComp_select_mat_noLabels, na.rm = T)
) / 2,
max(allByAllComp_select_mat_noLabels, na.rm = T)
),
c("#4575b4", "#ffffbf", "#d73027"))
#c("#ffeda0", "#feb24c", "#f03b20"))
topAnnoDat = tibble(name = colnames(allByAllComp_select_mat)) %>%
mutate(sampchrom = gsub("_v3", "", gsub("-.*", "", name))) %>%
mutate(sample = gsub("Dd2", "DD2", gsub("_.*", "", gsub("Pf", "", sampchrom)))) %>%
mutate(chrom = gsub(".*_", "", sampchrom)) %>%
left_join(metaByBioSample %>%
select(sample, secondaryRegion))
topAnno_df = topAnnoDat[,c("chrom", "secondaryRegion")] %>%
rename(continent = secondaryRegion) %>%
mutate(continent = gsub("Netherlands", "AFRICA", continent) ) %>%
as.data.frame()
topAnnoColors = createColorListFromDf(topAnno_df)
topAnnoColors[["continent"]] = c("AFRICA" = "#E69F00",
"ASIA" = "#F0E442",
"OCEANIA" = "#F748A5",
"S_AMERICA" = "#0072B2",
"Netherlands"="black")
topAnnoColors[["chrom"]] = c("11" = "#008DF9", "13" = "#A40122")
topAnno = HeatmapAnnotation(
df = topAnno_df,
col = topAnnoColors,
show_legend = F,
gp = gpar(col = "black")
# the annotation direction doesn't appear to work right now
# annotation_legend_param = list(
# chrom = list(legend_direction = "horizontal", direction = "horizontal"),
# continent = list(legend_direction = "horizontal", direction = "horizontal")
# )
)
sideAnno = rowAnnotation(
df = topAnno_df,
col = topAnnoColors,
show_legend = F,
gp = gpar(col = "black")
# ,
# annotation_legend_param = list(
# chrom = list(legend_direction = "horizontal", direction = "horizontal"),
# continent = list(legend_direction = "horizontal", direction = "horizontal")
# )
)
colnames(allByAllComp_select_mat_noLabels) = gsub("3D7", "**3D7", colnames(allByAllComp_select_mat_noLabels))
rownames(allByAllComp_select_mat_noLabels) = gsub("3D7", "**3D7", rownames(allByAllComp_select_mat_noLabels))
hc = hclust(dist(allByAllComp_select_mat_noLabels))
allByAllComp_select_mat_heatmap = Heatmap(
allByAllComp_select_mat_noLabels,
rect_gp = gpar(type = "none"),
column_dend_side = "bottom",
cell_fun = function(j, i, x, y, w, h, fill) {
if (as.numeric(x) <= 1 - as.numeric(y) + 1e-6) {
grid.rect(x, y, w, h, gp = gpar(fill = fill, col = fill))
}
},
# cluster_rows = hc,
# cluster_columns = hc,
row_names_side = "left",
name = "%Identity",
col = col_fun,
bottom_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
show_column_dend = F,
show_heatmap_legend = F
# ,
# heatmap_legend_param = list(
# direction = "horizontal"
# )
)
chrom_lgd = Legend(labels = sort(unique(topAnno_df$chrom)), title = "chrom", legend_gp = gpar(fill = topAnnoColors[["chrom"]][sort(unique(topAnno_df$chrom))]))
continent_lgd = Legend(labels = sort(unique(topAnno_df$continent)), title = "continent", legend_gp = gpar(fill = topAnnoColors[["continent"]][sort(unique(topAnno_df$continent))]))
heatmap_lgd = Legend(col_fun = col_fun, title = "%Identity")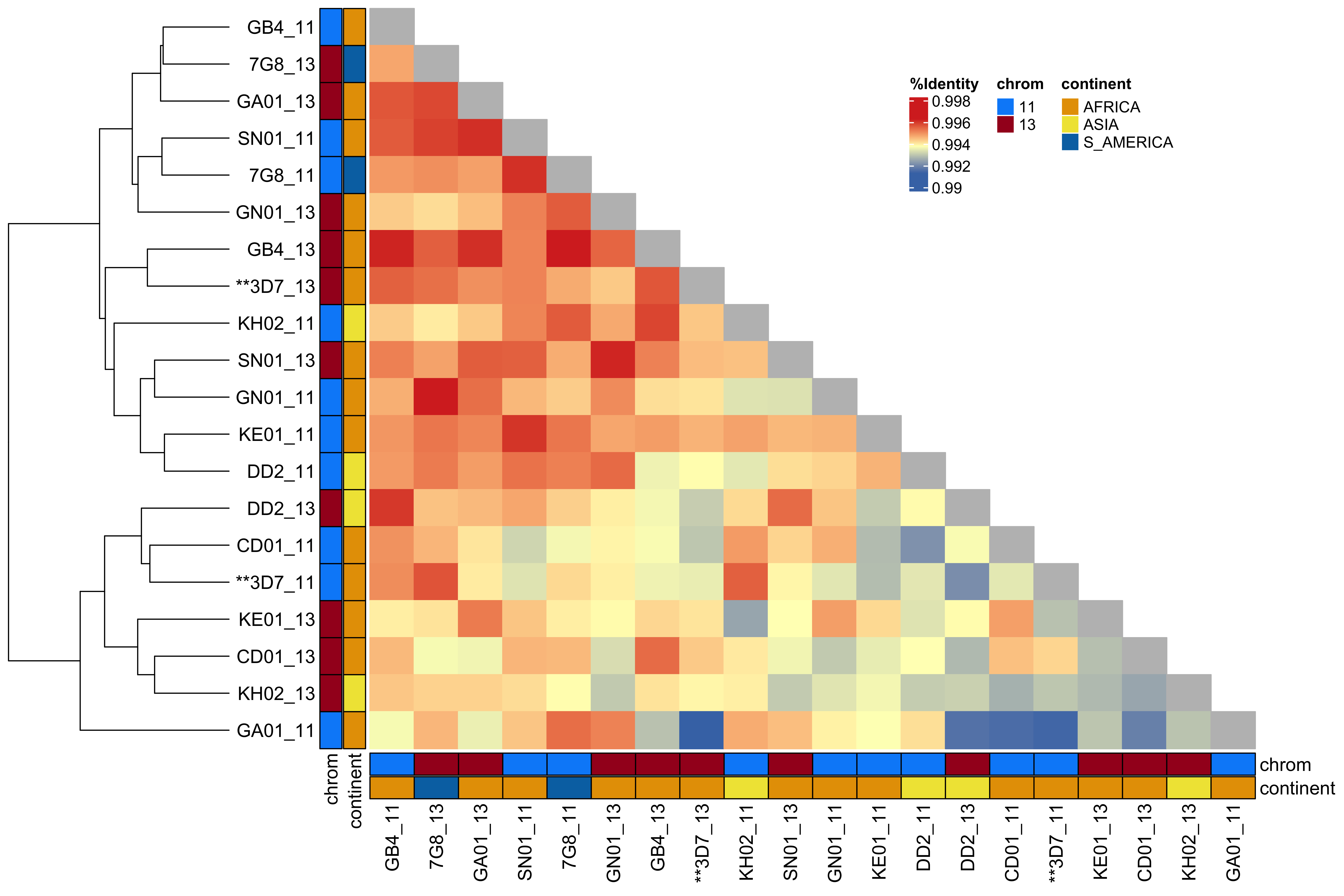
pdf("sharedRegion_pacbioAssemblies_percentIdentity.pdf", width = 7.5, height = 5, useDingbats = F)
draw(allByAllComp_select_mat_heatmap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "left")
draw(packLegend(heatmap_lgd,chrom_lgd,continent_lgd, direction = "horizontal"), x = unit(0.775, "npc"), y = unit(0.85, "npc"))
dev.off()quartz_off_screen
2 allByAllCompVarCount_select = allByAllComp %>%
select(OtherReadId, ReadId, totalDiffs) %>%
spread(ReadId, totalDiffs)
addRow = matrix(c(colnames(allByAllCompVarCount_select)[2], rep(NA, ncol(allByAllCompVarCount_select) - 1)), nrow = 1)
colnames(addRow) = colnames(allByAllCompVarCount_select)
addRow = as_tibble(addRow)
for(col in 2:ncol(addRow)){
addRow[,col] = as.numeric(addRow[,col])
}
allByAllCompVarCount_select = as_tibble(addRow) %>% bind_rows(allByAllCompVarCount_select)
allByAllCompVarCount_select_mat = as.matrix(allByAllCompVarCount_select[,2:ncol(allByAllCompVarCount_select)])
rownames(allByAllCompVarCount_select_mat) = allByAllCompVarCount_select$OtherReadId
allByAllCompVarCount_select_mat = as.matrix(as.dist(allByAllCompVarCount_select_mat))
allByAllCompVarCount_select_mat[0 == allByAllCompVarCount_select_mat] = NAlibrary(circlize)
meta = readr::read_tsv("../../meta/metadata//meta.tab.txt") %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
metaByBioSample = readr::read_tsv("../../meta/metadata/metaByBioSample.tab.txt") %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
allByAllCompVarCount_select_mat_noLabels = allByAllCompVarCount_select_mat
# rownames(allByAllCompVarCount_select_mat_noLabels) = NULL
# colnames(allByAllCompVarCount_select_mat_noLabels) = NULL
rownames(allByAllCompVarCount_select_mat_noLabels) = gsub("Dd2", "DD2", gsub("Pf", "", gsub("_v3", "", gsub("-.*", "", rownames(allByAllCompVarCount_select_mat_noLabels)))))
colnames(allByAllCompVarCount_select_mat_noLabels) = gsub("Dd2", "DD2", gsub("Pf", "", gsub("_v3", "", gsub("-.*", "", colnames(allByAllCompVarCount_select_mat_noLabels)))))
col_fun = colorRamp2(c(
min(allByAllCompVarCount_select_mat_noLabels, na.rm = T),
min(allByAllCompVarCount_select_mat_noLabels, na.rm = T) + (
max(allByAllCompVarCount_select_mat_noLabels, na.rm = T) - min(allByAllCompVarCount_select_mat_noLabels, na.rm = T)
) / 2,
max(allByAllCompVarCount_select_mat_noLabels, na.rm = T)
),
c("#006837", "#ffffbf", "#d73027"))
#c("#4575b4", "#ffffbf", "#d73027"))
#c("#ffeda0", "#feb24c", "#f03b20"))
topAnnoDat = tibble(name = colnames(allByAllCompVarCount_select_mat)) %>%
mutate(sampchrom = gsub("_v3", "", gsub("-.*", "", name))) %>%
mutate(sample = gsub("Dd2", "DD2", gsub("_.*", "", gsub("Pf", "", sampchrom)))) %>%
mutate(chrom = gsub(".*_", "", sampchrom)) %>%
left_join(metaByBioSample %>%
select(sample, secondaryRegion))
topAnno_df = topAnnoDat[,c("chrom", "secondaryRegion")] %>%
rename(continent = secondaryRegion) %>%
mutate(continent = gsub("Netherlands", "AFRICA", continent) ) %>%
as.data.frame()
topAnnoColors = createColorListFromDf(topAnno_df)
topAnnoColors[["continent"]] = c("AFRICA" = "#E69F00",
"ASIA" = "#F0E442",
"OCEANIA" = "#F748A5",
"S_AMERICA" = "#0072B2",
"Netherlands"="black")
topAnnoColors[["chrom"]] = c("11" = "#008DF9", "13" = "#A40122")
topAnno = HeatmapAnnotation(
df = topAnno_df,
col = topAnnoColors,
show_legend = F,
gp = gpar(col = "black")
# the annotation direction doesn't appear to work right now
# annotation_legend_param = list(
# chrom = list(legend_direction = "horizontal", direction = "horizontal"),
# continent = list(legend_direction = "horizontal", direction = "horizontal")
# )
)
sideAnno = rowAnnotation(
df = topAnno_df,
col = topAnnoColors,
show_legend = F,
gp = gpar(col = "black")
# ,
# annotation_legend_param = list(
# chrom = list(legend_direction = "horizontal", direction = "horizontal"),
# continent = list(legend_direction = "horizontal", direction = "horizontal")
# )
)
colnames(allByAllCompVarCount_select_mat_noLabels) = gsub("3D7", "**3D7", colnames(allByAllCompVarCount_select_mat_noLabels))
rownames(allByAllCompVarCount_select_mat_noLabels) = gsub("3D7", "**3D7", rownames(allByAllCompVarCount_select_mat_noLabels))
hc = hclust(dist(allByAllCompVarCount_select_mat_noLabels))
allByAllCompVarCount_select_mat_heatmap = Heatmap(
allByAllCompVarCount_select_mat_noLabels,
rect_gp = gpar(type = "none"),
column_dend_side = "bottom",
cell_fun = function(j, i, x, y, w, h, fill) {
if (as.numeric(x) <= 1 - as.numeric(y) + 1e-6) {
grid.rect(x, y, w, h, gp = gpar(fill = fill, col = fill))
}
},
# cluster_rows = hc,
# cluster_columns = hc,
row_names_side = "left",
name = "Number of \nVariants",
col = col_fun,
bottom_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
show_column_dend = F,
show_heatmap_legend = F,
na_col = "grey80",
# ,
# heatmap_legend_param = list(
# direction = "horizontal"
# )
)
chrom_lgd = Legend(labels = sort(unique(topAnno_df$chrom)), title = "chrom", legend_gp = gpar(fill = topAnnoColors[["chrom"]][sort(unique(topAnno_df$chrom))]))
continent_lgd = Legend(labels = sort(unique(topAnno_df$continent)), title = "continent", legend_gp = gpar(fill = topAnnoColors[["continent"]][sort(unique(topAnno_df$continent))]))
heatmap_lgd = Legend(col_fun = col_fun, title = "Number of \nVariants")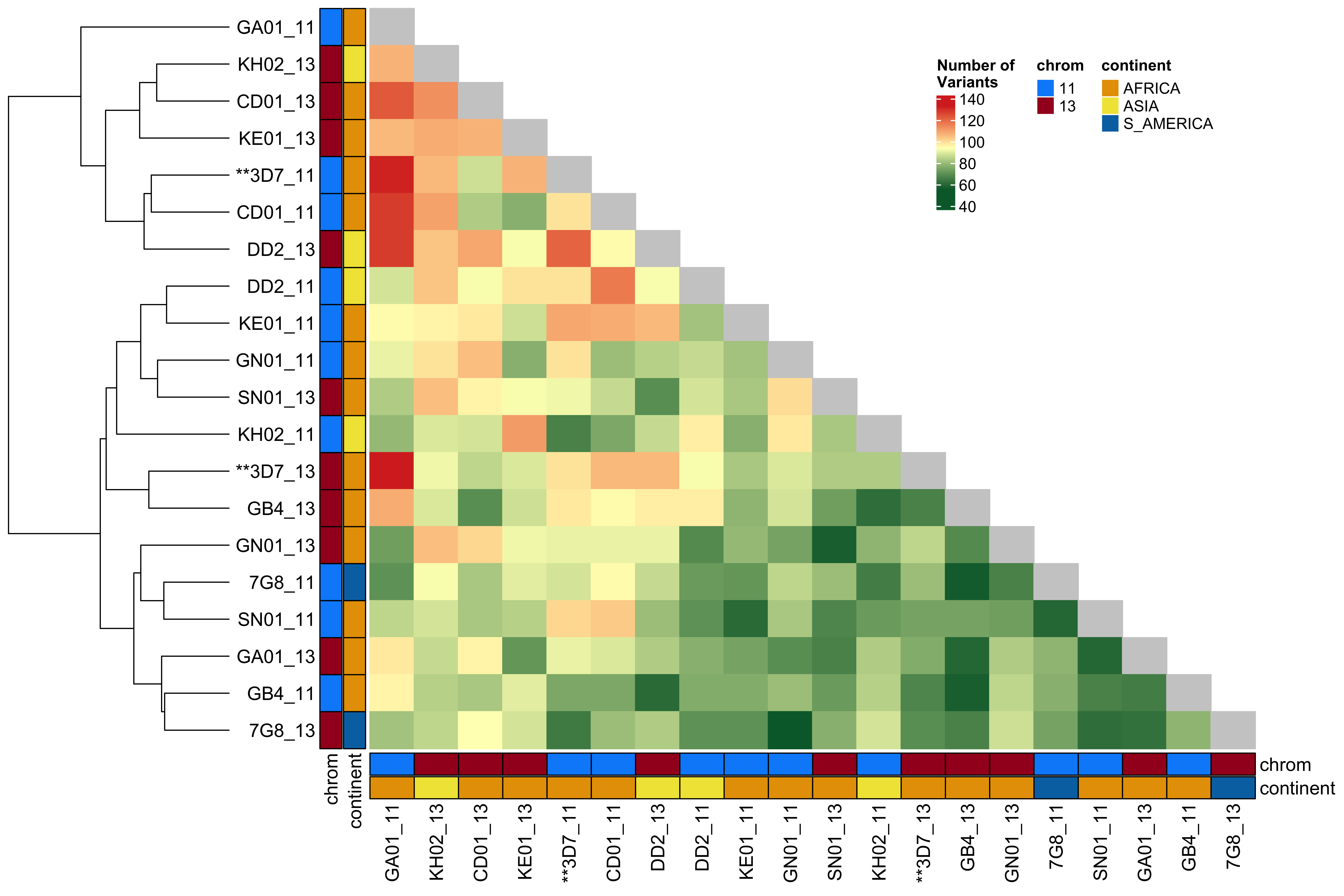
pdf("sharedRegion_pacbioAssemblies_varCount.pdf", width = 7.5, height = 5, useDingbats = F)
draw(allByAllCompVarCount_select_mat_heatmap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "left")
draw(packLegend(heatmap_lgd,chrom_lgd,continent_lgd, direction = "horizontal"), x = unit(0.80, "npc"), y = unit(0.85, "npc"))
dev.off()quartz_off_screen
2 allByAllComp_select_mat_noLabels_mod = allByAllComp_select_mat_noLabels
allByAllComp_select_mat_noLabels_mod = allByAllComp_select_mat_noLabels_mod - min(allByAllComp_select_mat_noLabels_mod, na.rm = T)
allByAllComp_select_mat_noLabels_mod[is.na(allByAllComp_select_mat_noLabels_mod)] = 0
Heatmap3D(allByAllComp_select_mat_noLabels_mod - min(allByAllComp_select_mat_noLabels_mod))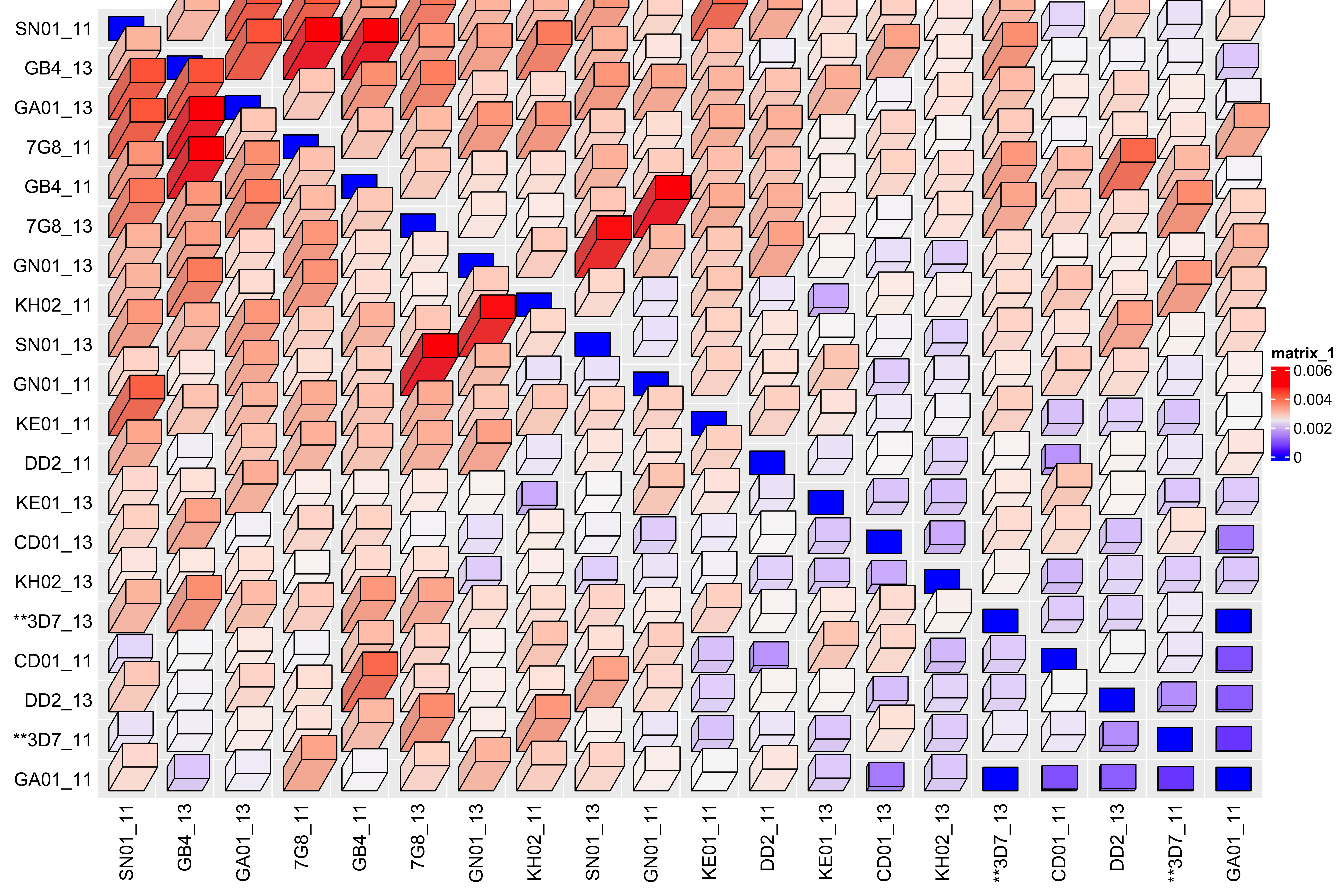
Comparison of the chromosomes vs between chromosomes, for chromosome 11 and 13 they are just as closely related to the same chromosome as between the chromosomes, unlike the chr05 and chr07 chromosomes rRNA loci where the same chromosomes are more closely related than between chromosomes. Could be suggestive that chr11 and chr13 are inter-combining more than chr05 and chr07
allByAllComp_mod = allByAllComp %>%
group_by(ReadId, OtherReadId) %>%
mutate(read1_chrom = ifelse(grepl("_11", ReadId), "chrom11", "chrom13"))%>%
mutate(read2_chrom = ifelse(grepl("_11", OtherReadId), "chrom11", "chrom13")) %>%
mutate(comparisonCategory = paste0(sort(c(read1_chrom, read2_chrom)), collapse = "--") ) %>%
ungroup()
ggplot(allByAllComp_mod) +
geom_boxplot(aes(x = comparisonCategory, y = perId)) +
sofonias_theme +
#scale_y_continuous(limits = c(min(allByAllComp_mod$perId), 1)) +
scale_y_continuous(limits = c(0.990, 1))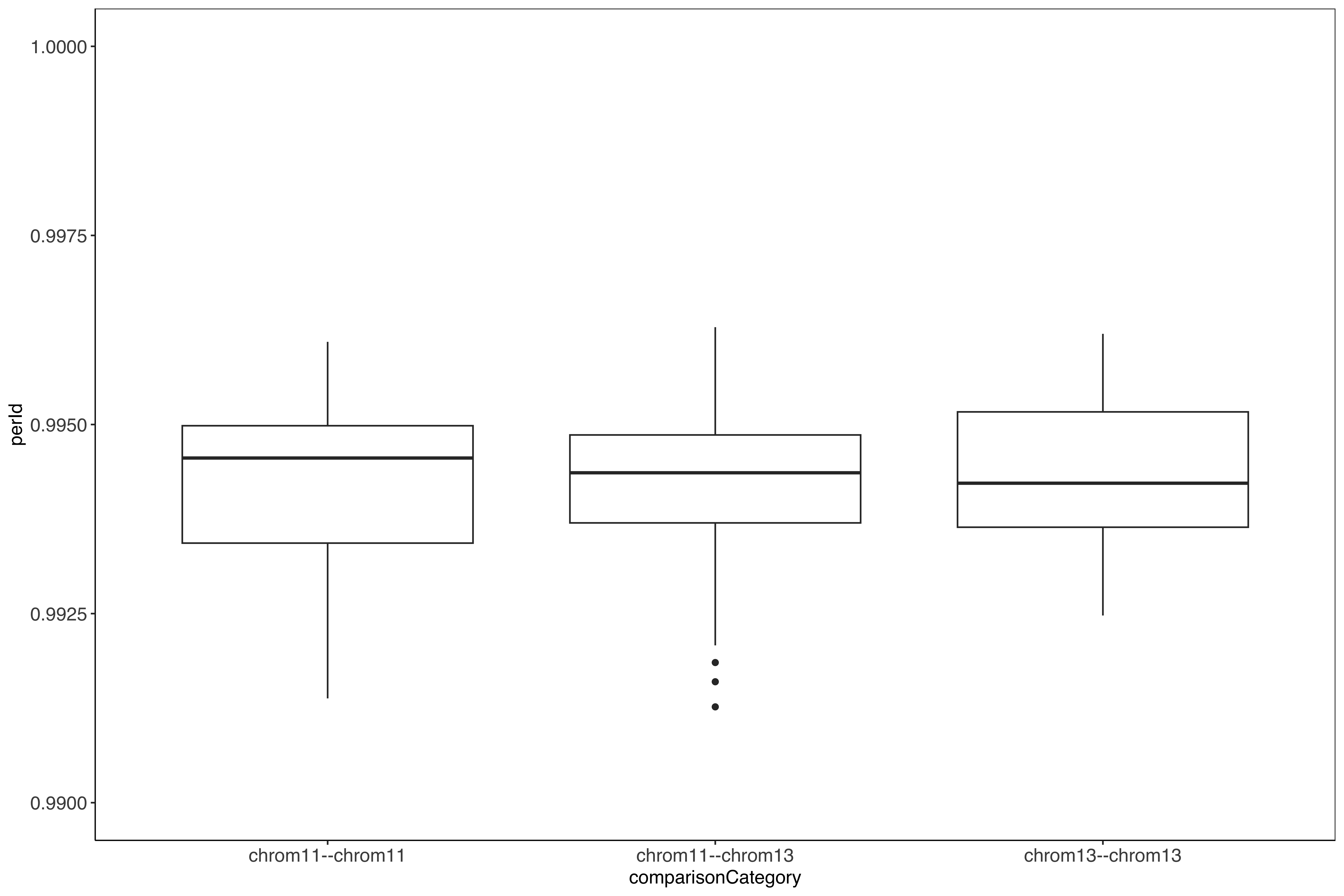
outBed = tibble(
`#chrom` = c("Pf3D7_11_v3", "Pf3D7_13_v3"),
start = c(1918029, 2792022),
end = c(1933390, 2807397),
name = c("Pf3D7_11_v3-1918029-1918124--Pf3D7_11_v3-1933291-1933390__Pf3D7_13_v3-2792022-2792117--Pf3D7_13_v3-2807298-2807397",
"Pf3D7_11_v3-1918029-1918124--Pf3D7_11_v3-1933291-1933390__Pf3D7_13_v3-2792022-2792117--Pf3D7_13_v3-2807298-2807397")
)
outBed = outBed %>%
mutate(len = end -start) %>%
mutate(strand = "+")
create_dt(outBed)write_tsv(outBed, "investigatingChrom11Chrom13/shared_11_13_region.bed")---
title: "Characterizing Duplicated Region"
---
```{r setup, echo=FALSE, message=FALSE}
source("../../common.R")
```
## Investigating shared region within pacbio assemblies[@Otto2018-bb]
```{bash, eval = F}
for x in /tank/data/genomes/plasmodium/genomes/pf/genomes/*.fasta; do elucidator splitFile --fasta ${x} --overWrite --trimAtWhiteSpace ; done;
```
These strains have complete chromosome 11s and 13s
Pf3D7
Pf7G8
PfCD01
PfDd2
PfGA01
PfGB4
PfGN01
PfKE01
PfKH02
PfSN01
```{r}
strains = c("Pf3D7","Pf7G8","PfCD01","PfDd2","PfGA01","PfGB4","PfGN01","PfKE01","PfKH02","PfSN01")
cat(strains, sep = "\n", file = "stableStrains.txt")
strainsDf = tibble(strain = strains)
strainsDf = strainsDf %>%
mutate(chrom11 = paste0(strain, "_11*.fasta")) %>%
mutate(chrom13 = paste0(strain, "_13*.fasta"))
mvCmd = paste0("mv ", paste0(strainsDf$chrom11, collapse = " "), " ", paste0(strainsDf$chrom13, collapse = " "), " combinedChrom11_13/")
cat(mvCmd)
```
```{bash, eval = F}
elucidator profileSharedKmerBlocks --seq Pf3D7_11_v3.fasta --dout chroms11_13_klen31 --kLen 31 --seqsDir ./ --fasta Pf3D7_11_v3.fasta --overWriteDir
cd chroms11_13_klen31
elucidator bedCoordSort --bed perfect.bed | elucidator extendBedRegions --left 5 --right 5 --bed STDIN | bedtools merge | elucidator bed3ToBed6 --bed STDIN | elucidator filterBedRecordsByLength --bed STDIN --minLen 65 | elucidator bedGetIntersectingGenesInGff --gff /work/nhathawa/genomes/pfGenomes_updated/info//gff/Pf3D7.gff --extraAttributes description --overWrite --bed STDIN --out perfectlyShared_minlen65.bed
elucidator getFastaWithBed --twoBit /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.2bit --overWrite --bed perfectlyShared_minlen65.bed --out perfectlyShared_minlen65
elucidator extractRefSeqsFromGenomes --genomeDir /tank/data/genomes/plasmodium/genomes/pf/genomes/ --gffDir /tank/data/genomes/plasmodium/genomes/pf/info/gff/ --primaryGenome Pf3D7 --bed perfectlyShared_minlen65.bed --outputDir extractedRefSeqs --identity 95 --coverage 95 --numThreads 15
```
```{r}
perfectlyShared_minlen65_regions = readr::read_tsv("combinedChrom11_13/chroms11_13_klen31/perfectlyShared_minlen65.bed", col_names = F)
perfectlyShared_minlen65_regions = perfectlyShared_minlen65_regions %>%
mutate(X4 = paste0(X4, "-for"))
allRegions = tibble()
for(region in perfectlyShared_minlen65_regions$X4){
for(strain in strains){
fnp= paste0("combinedChrom11_13/chroms11_13_klen31/extractedRefSeqs/", region, "/beds/", strain, "_region.bed")
if(file.exists(fnp)){
currentRegion = readr::read_tsv(fnp, col_names = F, show_col_types = F)
currentRegion$X4 = region
currentRegion$strain = strain
allRegions = bind_rows(
allRegions, currentRegion
)
}
}
}
allRegions_sum = allRegions %>%
group_by(strain, X1) %>%
summarise(start = min(X2),
end = max(X3)) %>%
filter(grepl("_1[13]", X1)) %>%
rename(`#chrom` = X1) %>%
mutate(start = start - 5000) %>%
mutate(end = end + 5000) %>%
mutate(name = paste0(`#chrom`, "-", start, "-", end),
len = end - start,
strand = "+")
for(strainName in strains){
allRegions_sum_byStrain = allRegions_sum %>%
filter(strain == strainName)
allRegions_sum_byStrain = allRegions_sum_byStrain %>%
group_by() %>%
select(-strain)
write_tsv(allRegions_sum_byStrain, paste0("combinedChrom11_13/chroms11_13_regions/", strainName, ".bed"))
}
allRegions_sum_exact = allRegions %>%
group_by(strain, X1) %>%
summarise(start = min(X2),
end = max(X3)) %>%
filter(grepl("_1[13]", X1)) %>%
rename(`#chrom` = X1) %>%
mutate(start = start) %>%
mutate(end = end) %>%
mutate(name = paste0(`#chrom`, "-", start, "-", end),
len = end - start,
strand = "+")
for(strainName in strains){
allRegions_sum_byStrain = allRegions_sum_exact %>%
filter(strain == strainName)
allRegions_sum_byStrain = allRegions_sum_byStrain %>%
group_by() %>%
select(-strain)
write_tsv(allRegions_sum_byStrain, paste0("combinedChrom11_13/chroms11_13_regions_exact/", strainName, ".bed"))
}
```
```{bash, eval = F}
cd chroms11_13_regions_exact
for x in *.bed; do elucidator getFastaWithBed --bed $x --twoBit /tank/data/genomes/plasmodium/genomes/pf/genomes/${x%%.bed}.2bit --overWrite --out ${x%%.bed}.fasta ; done;
mkdir sequences
cd sequences
for x in ../*.fasta; do elucidator splitFile --fasta $x ; done;
elucidator profileSharedKmerBlocks --seq Pf3D7_11_v3-1913023-1938396.fasta --dout chroms11_13_klen31 --kLen 31 --seqsDir ./ --fasta Pf3D7_11_v3-1913023-1938396.fasta --overWriteDir
```
```{bash, eval = F}
cd chroms11_13_regions
mkdir allSequences
for x in Pf*.bed; do elucidator getFastaWithBed --bed $x --twoBit /tank/data/genomes/plasmodium/genomes/pf/genomes/${x%%.bed}.2bit --overWrite --out ${x%%.bed}.fasta ; done;
cat *.fasta > allSequences/all.fasta
cat Pf*.bed > all.bed
cd allSequences
muscle -in all.fasta -out aln_all.fasta
elucidator trimToLen --fasta aln_all.fasta --length 21945 --overWrite --out trimmed_to_21945_aln_all.fasta
elucidator trimFront --fasta trimmed_to_21945_aln_all.fasta --forwardBases 21845 --overWrite --out trimmedFrom_21845_trimmed_to_21945_aln_all.fasta
elucidator sortReads --fasta trimmedFrom_21845_trimmed_to_21945_aln_all.fasta --removeGaps --overWrite
elucidator determineRegionLastz --individual --fasta sorted_trimmedFrom_21845_trimmed_to_21945_aln_all.fasta --genome /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.2bit -out sorted_trimmedFrom_21845_trimmed_to_21945_aln_all.bed
elucidator trimToLen --fasta aln_all.fasta --length 6051 --overWrite --out trimmed_to_6051_aln_all.fasta
elucidator trimFront --fasta trimmed_to_6051_aln_all.fasta --forwardBases 5951 --overWrite --out trimmedFrom_5951_trimmed_to_6051_aln_all.fasta
elucidator sortReads --fasta trimmedFrom_5951_trimmed_to_6051_aln_all.fasta --removeGaps --overWrite
elucidator determineRegionLastz --individual --fasta sorted_trimmedFrom_5951_trimmed_to_6051_aln_all.fasta --genome /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.2bit --overWrite --out sorted_trimmedFrom_5951_trimmed_to_6051_aln_all.bed
elucidatorlab getMalnConservedness --fasta aln_all.fasta --blockSize 10 --overWrite --out aln_all_conserved.tab.txt
elucidatorlab getAlnPosToRealPosTable --fasta aln_all.fasta --overWrite --out aln_all_position_table.tab.txt
```
## Beginning of the section
Pf3D7_11_v3-1918029-1918124--Pf3D7_13_v3-2792022-2792117
::: column-screen-inset-shaded
{width=70% fig-align="center"}
:::
## Middle
Variation in the middle, does not appear to be chromosome specific (though could have been a difference in assembly)
::: column-screen-inset-shaded
{width=70% fig-align="center"}
:::
## End of the section
Pf3D7_11_v3-1933291-1933390--Pf3D7_13_v3-2807298-2807397
::: column-screen-inset-shaded
{width=70% fig-align="center"}
:::
The region is ~15,360 kp long, so could be hard to get through even with long reads
## Conserved score
Plotting the percent conserved per position across the region including the before and after shared segment, all strains appear to start and end at the same position
```{r}
conservedPosTab = readr::read_tsv("combinedChrom11_13/chroms11_13_regions/allSequences/aln_all_position_table.tab.txt") %>%
filter("Pf3D7_11_v3-1913023-1938396" == name) %>%
rename(pos = alignPosition)
conservedMulti = readr::read_tsv("combinedChrom11_13/chroms11_13_regions/allSequences/aln_all_conserved.tab.txt")
conservedMulti_max = conservedMulti %>%
group_by(pos) %>%
filter(frac == max(frac)) %>%
filter("----------" != block ) %>%
left_join(conservedPosTab) %>%
filter(!is.na(name)) %>%
filter()
blockSize = 30
conservedMulti_max_rounding = tibble(rawPos = 1:(nrow(conservedMulti_max) - blockSize),
realPosition = numeric(nrow(conservedMulti_max) - blockSize),
rounded = numeric(nrow(conservedMulti_max) - blockSize))
for(row in 1:(nrow(conservedMulti_max) - blockSize)){
round = mean(conservedMulti_max$frac[row:(row+blockSize)])
if(round > 1){
print(row)
print(blockSize)
print(conservedMulti_max$frac[row:(row+blockSize)])
print(round)
stop()
}
minPos = min(conservedMulti_max$realPosition[row:(row+blockSize)])
conservedMulti_max_rounding$realPosition[row] = minPos
conservedMulti_max_rounding$rounded[row] = round
}
conservedMulti_max_rounding_filt = conservedMulti_max_rounding %>%
group_by(realPosition) %>%
mutate(id = row_number()) %>%
filter(id == 1) %>%
group_by()
```
```{r}
#| fig-column: screen-inset-shaded
ggplot(conservedMulti_max_rounding_filt) +
geom_bar(aes(x = realPosition, y = rounded), stat = "identity")+
sofonias_theme
```
```{bash, eval = F}
cd chroms11_13_regions_exact
cat *.fasta > all.fasta
elucidator compareAllByAll --fasta all.fasta --verbose --numThreads 12
elucidator createSharedSubSegmentsFromRefSeqs --fasta out.fasta --minimumKlen 12 --dout sharedSubSeqs_occurence6 --filterRegionsWithinRegions --refSeqName Pf3D7_11_v3-1918023-1933396 --overWriteDir --genome /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.fasta --minSubRegionLen 150 --maxSubRegionLen 300 --minBlockRegionLen 30 --doNotCollapseLowFreqNodes --correctionOccurenceCutOff 6 --includeFromSubRegionSize 30
muscle -in all.fasta -out aln_all.fasta
FastTree -nt aln_all.fasta > aln_all.nwk
elucidatorlab getMalnConservedness --fasta aln_all.fasta --blockSize 10 --overWrite --out aln_all_conserved.tab.txt
elucidatorlab getAlnPosToRealPosTable --fasta aln_all.fasta --overWrite --out aln_all_position_table.tab.txt
elucidator multipleAlnProteinToPcaInput --fasta aln_all.fasta --overWrite --out pcaMat_aln_all.fasta
elucidator runTRF --fasta all.fasta --overWriteDir --dout all_trfOutput --supplement --genomicLocation ../all.bed
elucidator bedFilterRegionsCompletelyInOther --bed all_trfOutput/genomic_combined.bed --intersectWithBed all_trfOutput/genomic_combined.bed | elucidator bedAddSmartIDForPlotting --bed STDIN --out all_trfOutput/filtered_genomic_combined.bed --overWrite
elucidator countBasesPerPosition --fasta aln_all.fasta --overWrite --out aln_all_baseCounts.tab.txt
#elucidator createSharedSubSegmentsFromRefSeqs --fasta out.fasta --minimumKlen 12 --dout sharedSubSeqs_occurence6 --filterRegionsWithinRegions --refSeqName Pf3D7_11_v3-1918023-1933396 --overWriteDir --genome /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.fasta --minSubRegionLen 150 --maxSubRegionLen 300 --minBlockRegionLen 30 --doNotCollapseLowFreqNodes --correctionOccurenceCutOff 6 --includeFromSubRegionSize 30
#cd sharedSubSeqs_occurence6/sharedLocs
#elucidator bedGetIntersectingGenesInGff --gff /tank/data/genomes/plasmodium/genomes/pf/info/gff/Pf3D7.gff --extraAttributes description --overWrite --bed 0_ref_sharedLocs_genomic.bed --out 0_ref_sharedLocs_genomic_withGeneInfo.bed
```
## Comparing chromsome 11 and 13 segments
The chromosome 11 and 13 regions appear to be extremely closely related to each other and there is no clear variation specific to chromosome 11 or 13
```{r}
library(ggtree)
tree = ape::read.tree("combinedChrom11_13/chroms11_13_regions_exact/sequences/aln_all.nwk")
tree$tip.label = gsub("_v3", "", gsub("-.*", "", tree$tip.label))
tipDf = tibble(seq = tree$tip.label) %>%
mutate(chrom = ifelse(grepl("_11", seq), "chrom11", "chrom13"))
tipDf_chrom11 = tipDf %>%
filter(chrom == "chrom11")
tipDf_chrom13 = tipDf %>%
filter(chrom == "chrom13")
groups = list(chrom11 = tipDf_chrom11$seq,
chrom13 = tipDf_chrom13$seq)
tree = groupOTU(tree, groups)
sharedRegion_pacbioAssembliesPlot = ggtree::ggtree(tree, show.legend = FALSE) +
hexpand(.1) +
geom_tiplab(aes(color = group), show.legend = FALSE) +
scale_color_manual(values = c("#005AC8", "#AA0A3C"))
```
```{r}
#| fig-column: screen-inset-shaded
#| fig-height: 5
#| fig-width: 10
print(sharedRegion_pacbioAssembliesPlot)
```
```{r}
pdf("sharedRegion_pacbioAssemblies.pdf", width = 7.5, height = 7.5/2, useDingbats = F)
print(sharedRegion_pacbioAssembliesPlot)
dev.off()
```
### Comparing Percent Identity
All segments between strains are more >99% related to each other
```{r}
allByAllComp = readr::read_tsv("combinedChrom11_13/chroms11_13_regions_exact/sequences/out.tab.txt")
```
```{r, eval=FALSE, echo=FALSE}
#| fig-column: screen-inset-shaded
ggplot(allByAllComp) +
geom_tile(aes(x = OtherReadId , y = ReadId, fill = perId)) +
geom_text(aes(x = OtherReadId , y = ReadId, label = round(perId*100,2))) +
sofonias_theme_xRotate +
scale_y_discrete(position = "right") +
scale_fill_gradient2(limits = c(min(allByAllComp$perId),1), midpoint = min(allByAllComp$perId) + (max(allByAllComp$perId) - min(allByAllComp$perId))/2 )
```
```{r}
allByAllComp_select = allByAllComp %>%
select(OtherReadId, ReadId, perId) %>%
spread(ReadId, perId)
addRow = matrix(c(colnames(allByAllComp_select)[2], rep(NA, ncol(allByAllComp_select) - 1)), nrow = 1)
colnames(addRow) = colnames(allByAllComp_select)
addRow = as_tibble(addRow)
for(col in 2:ncol(addRow)){
addRow[,col] = as.numeric(addRow[,col])
}
allByAllComp_select = as_tibble(addRow) %>% bind_rows(allByAllComp_select)
allByAllComp_select_mat = as.matrix(allByAllComp_select[,2:ncol(allByAllComp_select)])
rownames(allByAllComp_select_mat) = allByAllComp_select$OtherReadId
allByAllComp_select_mat = as.matrix(as.dist(allByAllComp_select_mat))
allByAllComp_select_mat[0 == allByAllComp_select_mat] = NA
```
```{r}
library(circlize)
meta = readr::read_tsv("../../meta/metadata/meta.tab.txt") %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
metaByBioSample = readr::read_tsv("../../meta/metadata/metaByBioSample.tab.txt") %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
allByAllComp_select_mat_noLabels = allByAllComp_select_mat
# rownames(allByAllComp_select_mat_noLabels) = NULL
# colnames(allByAllComp_select_mat_noLabels) = NULL
rownames(allByAllComp_select_mat_noLabels) = gsub("Dd2", "DD2", gsub("Pf", "", gsub("_v3", "", gsub("-.*", "", rownames(allByAllComp_select_mat_noLabels)))))
colnames(allByAllComp_select_mat_noLabels) = gsub("Dd2", "DD2", gsub("Pf", "", gsub("_v3", "", gsub("-.*", "", colnames(allByAllComp_select_mat_noLabels)))))
col_fun = colorRamp2(c(
min(allByAllComp_select_mat_noLabels, na.rm = T),
min(allByAllComp_select_mat_noLabels, na.rm = T) + (
max(allByAllComp_select_mat_noLabels, na.rm = T) - min(allByAllComp_select_mat_noLabels, na.rm = T)
) / 2,
max(allByAllComp_select_mat_noLabels, na.rm = T)
),
c("#4575b4", "#ffffbf", "#d73027"))
#c("#ffeda0", "#feb24c", "#f03b20"))
topAnnoDat = tibble(name = colnames(allByAllComp_select_mat)) %>%
mutate(sampchrom = gsub("_v3", "", gsub("-.*", "", name))) %>%
mutate(sample = gsub("Dd2", "DD2", gsub("_.*", "", gsub("Pf", "", sampchrom)))) %>%
mutate(chrom = gsub(".*_", "", sampchrom)) %>%
left_join(metaByBioSample %>%
select(sample, secondaryRegion))
topAnno_df = topAnnoDat[,c("chrom", "secondaryRegion")] %>%
rename(continent = secondaryRegion) %>%
mutate(continent = gsub("Netherlands", "AFRICA", continent) ) %>%
as.data.frame()
topAnnoColors = createColorListFromDf(topAnno_df)
topAnnoColors[["continent"]] = c("AFRICA" = "#E69F00",
"ASIA" = "#F0E442",
"OCEANIA" = "#F748A5",
"S_AMERICA" = "#0072B2",
"Netherlands"="black")
topAnnoColors[["chrom"]] = c("11" = "#008DF9", "13" = "#A40122")
topAnno = HeatmapAnnotation(
df = topAnno_df,
col = topAnnoColors,
show_legend = F,
gp = gpar(col = "black")
# the annotation direction doesn't appear to work right now
# annotation_legend_param = list(
# chrom = list(legend_direction = "horizontal", direction = "horizontal"),
# continent = list(legend_direction = "horizontal", direction = "horizontal")
# )
)
sideAnno = rowAnnotation(
df = topAnno_df,
col = topAnnoColors,
show_legend = F,
gp = gpar(col = "black")
# ,
# annotation_legend_param = list(
# chrom = list(legend_direction = "horizontal", direction = "horizontal"),
# continent = list(legend_direction = "horizontal", direction = "horizontal")
# )
)
colnames(allByAllComp_select_mat_noLabels) = gsub("3D7", "**3D7", colnames(allByAllComp_select_mat_noLabels))
rownames(allByAllComp_select_mat_noLabels) = gsub("3D7", "**3D7", rownames(allByAllComp_select_mat_noLabels))
hc = hclust(dist(allByAllComp_select_mat_noLabels))
allByAllComp_select_mat_heatmap = Heatmap(
allByAllComp_select_mat_noLabels,
rect_gp = gpar(type = "none"),
column_dend_side = "bottom",
cell_fun = function(j, i, x, y, w, h, fill) {
if (as.numeric(x) <= 1 - as.numeric(y) + 1e-6) {
grid.rect(x, y, w, h, gp = gpar(fill = fill, col = fill))
}
},
# cluster_rows = hc,
# cluster_columns = hc,
row_names_side = "left",
name = "%Identity",
col = col_fun,
bottom_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
show_column_dend = F,
show_heatmap_legend = F
# ,
# heatmap_legend_param = list(
# direction = "horizontal"
# )
)
chrom_lgd = Legend(labels = sort(unique(topAnno_df$chrom)), title = "chrom", legend_gp = gpar(fill = topAnnoColors[["chrom"]][sort(unique(topAnno_df$chrom))]))
continent_lgd = Legend(labels = sort(unique(topAnno_df$continent)), title = "continent", legend_gp = gpar(fill = topAnnoColors[["continent"]][sort(unique(topAnno_df$continent))]))
heatmap_lgd = Legend(col_fun = col_fun, title = "%Identity")
```
```{r}
#| fig-column: screen-inset-shaded
draw(allByAllComp_select_mat_heatmap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "left")
draw(packLegend(heatmap_lgd,chrom_lgd,continent_lgd, direction = "horizontal"), x = unit(0.775, "npc"), y = unit(0.85, "npc"))
```
```{r}
pdf("sharedRegion_pacbioAssemblies_percentIdentity.pdf", width = 7.5, height = 5, useDingbats = F)
draw(allByAllComp_select_mat_heatmap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "left")
draw(packLegend(heatmap_lgd,chrom_lgd,continent_lgd, direction = "horizontal"), x = unit(0.775, "npc"), y = unit(0.85, "npc"))
dev.off()
```
### Comparing Number of variants
```{r}
allByAllCompVarCount_select = allByAllComp %>%
select(OtherReadId, ReadId, totalDiffs) %>%
spread(ReadId, totalDiffs)
addRow = matrix(c(colnames(allByAllCompVarCount_select)[2], rep(NA, ncol(allByAllCompVarCount_select) - 1)), nrow = 1)
colnames(addRow) = colnames(allByAllCompVarCount_select)
addRow = as_tibble(addRow)
for(col in 2:ncol(addRow)){
addRow[,col] = as.numeric(addRow[,col])
}
allByAllCompVarCount_select = as_tibble(addRow) %>% bind_rows(allByAllCompVarCount_select)
allByAllCompVarCount_select_mat = as.matrix(allByAllCompVarCount_select[,2:ncol(allByAllCompVarCount_select)])
rownames(allByAllCompVarCount_select_mat) = allByAllCompVarCount_select$OtherReadId
allByAllCompVarCount_select_mat = as.matrix(as.dist(allByAllCompVarCount_select_mat))
allByAllCompVarCount_select_mat[0 == allByAllCompVarCount_select_mat] = NA
```
```{r}
library(circlize)
meta = readr::read_tsv("../../meta/metadata//meta.tab.txt") %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
metaByBioSample = readr::read_tsv("../../meta/metadata/metaByBioSample.tab.txt") %>%
mutate(country = gsub("South East Asia - East", "Cambodia", country))
allByAllCompVarCount_select_mat_noLabels = allByAllCompVarCount_select_mat
# rownames(allByAllCompVarCount_select_mat_noLabels) = NULL
# colnames(allByAllCompVarCount_select_mat_noLabels) = NULL
rownames(allByAllCompVarCount_select_mat_noLabels) = gsub("Dd2", "DD2", gsub("Pf", "", gsub("_v3", "", gsub("-.*", "", rownames(allByAllCompVarCount_select_mat_noLabels)))))
colnames(allByAllCompVarCount_select_mat_noLabels) = gsub("Dd2", "DD2", gsub("Pf", "", gsub("_v3", "", gsub("-.*", "", colnames(allByAllCompVarCount_select_mat_noLabels)))))
col_fun = colorRamp2(c(
min(allByAllCompVarCount_select_mat_noLabels, na.rm = T),
min(allByAllCompVarCount_select_mat_noLabels, na.rm = T) + (
max(allByAllCompVarCount_select_mat_noLabels, na.rm = T) - min(allByAllCompVarCount_select_mat_noLabels, na.rm = T)
) / 2,
max(allByAllCompVarCount_select_mat_noLabels, na.rm = T)
),
c("#006837", "#ffffbf", "#d73027"))
#c("#4575b4", "#ffffbf", "#d73027"))
#c("#ffeda0", "#feb24c", "#f03b20"))
topAnnoDat = tibble(name = colnames(allByAllCompVarCount_select_mat)) %>%
mutate(sampchrom = gsub("_v3", "", gsub("-.*", "", name))) %>%
mutate(sample = gsub("Dd2", "DD2", gsub("_.*", "", gsub("Pf", "", sampchrom)))) %>%
mutate(chrom = gsub(".*_", "", sampchrom)) %>%
left_join(metaByBioSample %>%
select(sample, secondaryRegion))
topAnno_df = topAnnoDat[,c("chrom", "secondaryRegion")] %>%
rename(continent = secondaryRegion) %>%
mutate(continent = gsub("Netherlands", "AFRICA", continent) ) %>%
as.data.frame()
topAnnoColors = createColorListFromDf(topAnno_df)
topAnnoColors[["continent"]] = c("AFRICA" = "#E69F00",
"ASIA" = "#F0E442",
"OCEANIA" = "#F748A5",
"S_AMERICA" = "#0072B2",
"Netherlands"="black")
topAnnoColors[["chrom"]] = c("11" = "#008DF9", "13" = "#A40122")
topAnno = HeatmapAnnotation(
df = topAnno_df,
col = topAnnoColors,
show_legend = F,
gp = gpar(col = "black")
# the annotation direction doesn't appear to work right now
# annotation_legend_param = list(
# chrom = list(legend_direction = "horizontal", direction = "horizontal"),
# continent = list(legend_direction = "horizontal", direction = "horizontal")
# )
)
sideAnno = rowAnnotation(
df = topAnno_df,
col = topAnnoColors,
show_legend = F,
gp = gpar(col = "black")
# ,
# annotation_legend_param = list(
# chrom = list(legend_direction = "horizontal", direction = "horizontal"),
# continent = list(legend_direction = "horizontal", direction = "horizontal")
# )
)
colnames(allByAllCompVarCount_select_mat_noLabels) = gsub("3D7", "**3D7", colnames(allByAllCompVarCount_select_mat_noLabels))
rownames(allByAllCompVarCount_select_mat_noLabels) = gsub("3D7", "**3D7", rownames(allByAllCompVarCount_select_mat_noLabels))
hc = hclust(dist(allByAllCompVarCount_select_mat_noLabels))
allByAllCompVarCount_select_mat_heatmap = Heatmap(
allByAllCompVarCount_select_mat_noLabels,
rect_gp = gpar(type = "none"),
column_dend_side = "bottom",
cell_fun = function(j, i, x, y, w, h, fill) {
if (as.numeric(x) <= 1 - as.numeric(y) + 1e-6) {
grid.rect(x, y, w, h, gp = gpar(fill = fill, col = fill))
}
},
# cluster_rows = hc,
# cluster_columns = hc,
row_names_side = "left",
name = "Number of \nVariants",
col = col_fun,
bottom_annotation = topAnno,
left_annotation = sideAnno,
row_dend_width = unit(5, "cm"),
column_dend_height = unit(5, "cm"),
show_column_dend = F,
show_heatmap_legend = F,
na_col = "grey80",
# ,
# heatmap_legend_param = list(
# direction = "horizontal"
# )
)
chrom_lgd = Legend(labels = sort(unique(topAnno_df$chrom)), title = "chrom", legend_gp = gpar(fill = topAnnoColors[["chrom"]][sort(unique(topAnno_df$chrom))]))
continent_lgd = Legend(labels = sort(unique(topAnno_df$continent)), title = "continent", legend_gp = gpar(fill = topAnnoColors[["continent"]][sort(unique(topAnno_df$continent))]))
heatmap_lgd = Legend(col_fun = col_fun, title = "Number of \nVariants")
```
```{r}
#| fig-column: screen-inset-shaded
draw(allByAllCompVarCount_select_mat_heatmap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "left")
draw(packLegend(heatmap_lgd,chrom_lgd,continent_lgd, direction = "horizontal"), x = unit(0.80, "npc"), y = unit(0.85, "npc"))
```
```{r}
pdf("sharedRegion_pacbioAssemblies_varCount.pdf", width = 7.5, height = 5, useDingbats = F)
draw(allByAllCompVarCount_select_mat_heatmap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "left")
draw(packLegend(heatmap_lgd,chrom_lgd,continent_lgd, direction = "horizontal"), x = unit(0.80, "npc"), y = unit(0.85, "npc"))
dev.off()
```
```{r, eval = F, echo=F}
library(gridExtra)
conservedInfo_between_11_and_13_sharedRegion = readr::read_tsv("../../MappingOutSurroundingRegions/conservedInfo_between_11_and_13_sharedRegion.tsv")
conservedInfo_current_plot = ggplot(conservedInfo_between_11_and_13_sharedRegion) +
geom_segment(
aes(
x = chrom1Pos_start,
xend = crhom1Pos_end - 1,
y = chrom2Pos,
yend = chrom2Pos + size - 1
),
linewidth = 2.5,
color = "black"
) +
geom_rect(
ymin = conservedInfo_between_11_and_13_sharedRegion$chrom2_section_start[1],
ymax = conservedInfo_between_11_and_13_sharedRegion$chrom2_section_start[1] + conservedInfo_between_11_and_13_sharedRegion$chrom2_section_len[1],
xmin = conservedInfo_between_11_and_13_sharedRegion$chrom1_section_start[1] - conservedInfo_between_11_and_13_sharedRegion$chrom1_section_len[1] * 0.1,
xmax = conservedInfo_between_11_and_13_sharedRegion$chrom1_section_start[1] - conservedInfo_between_11_and_13_sharedRegion$chrom1_section_len[1] * 0.001,
fill = "grey81") +
geom_rect(
ymax = conservedInfo_between_11_and_13_sharedRegion$chrom2_section_start[1] - conservedInfo_between_11_and_13_sharedRegion$chrom2_section_len[1] * 0.001,
ymin = conservedInfo_between_11_and_13_sharedRegion$chrom2_section_start[1] - conservedInfo_between_11_and_13_sharedRegion$chrom2_section_len[1] * 0.1,
xmin = conservedInfo_between_11_and_13_sharedRegion$chrom1_section_start[1],
xmax = conservedInfo_between_11_and_13_sharedRegion$chrom1_section_start[1] + conservedInfo_between_11_and_13_sharedRegion$chrom1_section_len[1],
fill = "grey81") +
geom_rect(data = pf3d7Genes_chrom1,
aes(xmin = col.1, xmax = col.2,
ymax = conservedInfo_between_11_and_13_sharedRegion$chrom2_section_start[1] - conservedInfo_between_11_and_13_sharedRegion$chrom2_section_len[1] * 0.001,
ymin = conservedInfo_between_11_and_13_sharedRegion$chrom2_section_start[1] - conservedInfo_between_11_and_13_sharedRegion$chrom2_section_len[1] * 0.1,
fill = description)) +
geom_rect(data = pf3d7Genes_chrom2,
aes(ymin = col.1, ymax = col.2,
xmax = conservedInfo_between_11_and_13_sharedRegion$chrom1_section_start[1] - conservedInfo_between_11_and_13_sharedRegion$chrom1_section_len[1] * 0.001,
xmin = conservedInfo_between_11_and_13_sharedRegion$chrom1_section_start[1] - conservedInfo_between_11_and_13_sharedRegion$chrom1_section_len[1] * 0.1,
fill = description)) +
labs(title = "",
x = "3D7 Chr13-2792021-2807295 (bp=15,274)",
y = "3D7 Chr11-1918028-1933288 (bp=15,260)",
fill = "") +
sofonias_theme +
coord_equal() +
# scale_fill_tableau() +
scale_fill_manual(values = c("#2271B2", "#F748A5", "#3DB7E9", "#D55E00")) +
guides(fill=guide_legend(ncol = 1,byrow=TRUE)) +
theme(legend.position = c(0.3, 0.7),
panel.border = element_blank(),
legend.background = element_blank(),
legend.box.background = element_rect(colour = "black"))
grobHeatmap = grid.grabExpr({
draw(allByAllComp_select_mat_heatmap, background = "transparent", merge_legend = TRUE, heatmap_legend_side = "bottom", annotation_legend_side = "left")
draw(packLegend(heatmap_lgd,chrom_lgd,continent_lgd, direction = "horizontal"), x = unit(0.775, "npc"), y = unit(0.85, "npc"))
})
grobGgplot = grid.grabExpr({
print(conservedInfo_current_plot)
})
gl = list(grobGgplot, grobHeatmap)
lay <- rbind(c(1,1,1,1,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA),
c(1,1,1,1,NA,NA,2,2,2,2,2,2,2,2),
c(1,1,1,1,NA,NA,2,2,2,2,2,2,2,2),
c(1,1,1,1,NA,NA,2,2,2,2,2,2,2,2),
c(1,1,1,1,NA,NA,2,2,2,2,2,2,2,2),
c(1,1,1,1,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA))
pdf("test.pdf", width = 10, height = 10, useDingbats = F)
grid.arrange(
grobs = gl,
layout_matrix = lay
)
dev.off()
```
```{r}
#| fig-column: screen-inset-shaded
allByAllComp_select_mat_noLabels_mod = allByAllComp_select_mat_noLabels
allByAllComp_select_mat_noLabels_mod = allByAllComp_select_mat_noLabels_mod - min(allByAllComp_select_mat_noLabels_mod, na.rm = T)
allByAllComp_select_mat_noLabels_mod[is.na(allByAllComp_select_mat_noLabels_mod)] = 0
Heatmap3D(allByAllComp_select_mat_noLabels_mod - min(allByAllComp_select_mat_noLabels_mod))
```
## Chromosome vs chromosome percent identity
Comparison of the chromosomes vs between chromosomes, for chromosome 11 and 13 they are just as closely related to the same chromosome as between the chromosomes, unlike the [chr05 and chr07 chromosomes rRNA loci](../sharedBetween05_and_07/characterizingSharingInPacbio.qmd#chromosome-vs-chromosome-percent-identity) where the same chromosomes are more closely related than between chromosomes. Could be suggestive that chr11 and chr13 are inter-combining more than chr05 and chr07
```{r}
allByAllComp_mod = allByAllComp %>%
group_by(ReadId, OtherReadId) %>%
mutate(read1_chrom = ifelse(grepl("_11", ReadId), "chrom11", "chrom13"))%>%
mutate(read2_chrom = ifelse(grepl("_11", OtherReadId), "chrom11", "chrom13")) %>%
mutate(comparisonCategory = paste0(sort(c(read1_chrom, read2_chrom)), collapse = "--") ) %>%
ungroup()
ggplot(allByAllComp_mod) +
geom_boxplot(aes(x = comparisonCategory, y = perId)) +
sofonias_theme +
#scale_y_continuous(limits = c(min(allByAllComp_mod$perId), 1)) +
scale_y_continuous(limits = c(0.990, 1))
```
# Writing out region
```{r}
outBed = tibble(
`#chrom` = c("Pf3D7_11_v3", "Pf3D7_13_v3"),
start = c(1918029, 2792022),
end = c(1933390, 2807397),
name = c("Pf3D7_11_v3-1918029-1918124--Pf3D7_11_v3-1933291-1933390__Pf3D7_13_v3-2792022-2792117--Pf3D7_13_v3-2807298-2807397",
"Pf3D7_11_v3-1918029-1918124--Pf3D7_11_v3-1933291-1933390__Pf3D7_13_v3-2792022-2792117--Pf3D7_13_v3-2807298-2807397")
)
outBed = outBed %>%
mutate(len = end -start) %>%
mutate(strand = "+")
create_dt(outBed)
write_tsv(outBed, "investigatingChrom11Chrom13/shared_11_13_region.bed")
```
<!-- ## Mapping out shared region -->
<!-- ```{r} -->
<!-- conservedPosTab = readr::read_tsv("combinedChrom11_13/chroms11_13_regions_exact/sequences/aln_all_position_table.tab.txt") %>% -->
<!-- separate(name, into = c("chrom", "start", "end"), convert = T, remove = F, sep = "-") %>% -->
<!-- mutate(realPosition = realPosition + start) -->
<!-- aln_all_baseCounts = readr::read_tsv("combinedChrom11_13/chroms11_13_regions_exact/sequences/aln_all_baseCounts.tab.txt") -->
<!-- aln_all_baseCounts_filt = aln_all_baseCounts %>% -->
<!-- filter(count > 0, fraction <1) %>% -->
<!-- group_by(position) %>% -->
<!-- filter(max(count) < 19) %>% -->
<!-- filter(!any(base == '-')) %>% -->
<!-- select(-meanReadLengthRounded) -->
<!-- conservedPosTab_Pf3D7_11_v3 = conservedPosTab %>% -->
<!-- filter(grepl("Pf3D7_11_v3", name)) %>% -->
<!-- rename(position = alignPosition) %>% -->
<!-- left_join(aln_all_baseCounts_filt)%>% -->
<!-- filter(!is.na(base)) -->
<!-- conservedPosTab_Pf3D7_13_v3 = conservedPosTab %>% -->
<!-- filter(grepl("Pf3D7_13_v3", name))%>% -->
<!-- rename(position = alignPosition) %>% -->
<!-- left_join(aln_all_baseCounts_filt) %>% -->
<!-- filter(!is.na(base)) -->
<!-- variableSitesInPacbioGenomes = bind_rows( -->
<!-- conservedPosTab_Pf3D7_11_v3, -->
<!-- conservedPosTab_Pf3D7_13_v3 -->
<!-- ) %>% -->
<!-- mutate(`#chrom` = chrom) -->
<!-- chroms = readr::read_tsv("../../MappingOutSurroundingRegions/surroundingRegionsMaterials/chromLengths/Pf3D7.txt", col_names = c("chrom", "length")) -->
<!-- chrom11_genes = readr::read_tsv("../../MappingOutSurroundingRegions/surroundingRegionsMaterials//endBeds/split_Pf3D7_chrom11_toEnd_genes.tab.txt", col_names = T) -->
<!-- chrom13_genes = readr::read_tsv("../../MappingOutSurroundingRegions/surroundingRegionsMaterials//endBeds/split_Pf3D7_chrom13_toEnd_genes.tab.txt", col_names = T) -->
<!-- allGenes = bind_rows(chrom11_genes, chrom13_genes) %>% -->
<!-- rename(chrom = col.0, start = col.1, end = col.2, name = col.3, length = col.4, strand = col.5) %>% -->
<!-- mutate(rawGeneDescription = description) %>% -->
<!-- mutate(gene = ifelse(grepl("PF3D7_0831800", description), "HRP II", "other")) %>% -->
<!-- mutate(gene = ifelse(grepl("PF3D7_1372200", description), "HRP III", gene)) %>% -->
<!-- mutate(description = gsub(" \\(SURFIN", "\\(SURFIN", description)) %>% -->
<!-- mutate(description = ifelse(grepl("Plasmodium exported protein", description), "Plasmodium exported protein (PHIST)", description)) %>% -->
<!-- mutate(description = ifelse("erythrocyte membrane protein 1 (PfEMP1), exon 2"==description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene", description)) %>% -->
<!-- mutate(description = ifelse("membrane associated erythrocyte binding-like protein"==description, "merozoite adhesive erythrocytic binding protein", description)) %>% -->
<!-- mutate(description = ifelse("membrane associated erythrocyte binding-likeprotein"==description, "merozoite adhesive erythrocytic binding protein", description)) %>% -->
<!-- mutate(description = ifelse("membrane associated histidine-rich protein 1"==description, "membrane associated histidine-rich protein", description)) %>% -->
<!-- mutate(description = gsub(",putative", ", putative", description)) %>% -->
<!-- mutate(description = gsub(",pseudogene", ", pseudogene", description))%>% -->
<!-- mutate(description = gsub("unknownfunction", "unknown function", description))%>% -->
<!-- mutate(description = gsub("conserved protein, unknown function", "conserved Plasmodium protein, unknown function", description)) %>% -->
<!-- mutate(description = gsub(" \\(SURFIN", "\\(SURFIN", description)) %>% -->
<!-- mutate(description = ifelse(grepl("Plasmodium exported protein", description), "Plasmodium exported protein (PHIST)", description)) %>% -->
<!-- mutate(description = ifelse("erythrocyte membrane protein 1 (PfEMP1), exon 2"==description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene", description)) %>% -->
<!-- mutate(description = ifelse("membrane associated histidine-rich protein 1"==description, "membrane associated histidine-rich protein", description))%>% -->
<!-- mutate(description = gsub(",pseudogene", ", pseudogene", description))%>% -->
<!-- mutate(description = gsub("unknownfunction", "unknown function", description))%>% -->
<!-- mutate(description = gsub(" \\(SURFIN", "\\(SURFIN", description)) %>% -->
<!-- mutate(description = ifelse(grepl("Plasmodium exported protein", description), "Plasmodium exported protein (PHIST)", description)) %>% -->
<!-- mutate(description = ifelse("erythrocyte membrane protein 1 (PfEMP1), exon 2"==description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene", description)) %>% -->
<!-- mutate(description = ifelse(grepl("sporozoite and liver stage tryptophan-rich protein, putative", description), "tryptophan/threonine-rich antigen", description))%>% -->
<!-- mutate(description = ifelse(grepl("CRA domain-containing protein, putative", description), "conserved Plasmodium protein, unknown function", description))%>% -->
<!-- mutate(description = gsub(",pseudogene", ", pseudogene", description)) %>% -->
<!-- mutate(description = gsub("surfaceantigen", "surface antigen", description)) %>% -->
<!-- mutate(description = gsub("Tetratricopeptide repeat, putative", "tetratricopeptide repeat protein, putative", description)) %>% -->
<!-- mutate(description = gsub("transmembraneprotein", "transmembrane protein", description)) %>% -->
<!-- mutate(description = ifelse(grepl("PfEMP1", description) & grepl("pseudogene", description), "erythrocyte membrane protein 1 (PfEMP1), pseudogene", description)) %>% -->
<!-- mutate(description = gsub("PIR protein", "stevor", description)) %>% -->
<!-- mutate(description = gsub("erythrocyte membrane protein 1-like", "erythrocyte membrane protein 1 (PfEMP1), pseudogene", description)) %>% -->
<!-- mutate(description = gsub("acidic terminal segments, variant surface antigen of PfEMP1, putative", "erythrocyte membrane protein 1 (PfEMP1), pseudogene", description))%>% -->
<!-- mutate(description = ifelse(grepl("CoA binding protein", description, ignore.case = T), "acyl-CoA binding protein", description)) %>% -->
<!-- mutate(description = ifelse(grepl("transfer RNA", description) | grepl("tRNA", description), "tRNA", description))%>% -->
<!-- mutate(description = ifelse(grepl("cytoadherence", description), "CLAG", description))%>% -->
<!-- mutate(description = ifelse(grepl("surface-associated interspersed protein", description), "SURFIN", description))%>% -->
<!-- mutate(description = ifelse(grepl("SURFIN", description), "SURFIN", description))%>% -->
<!-- mutate(description = ifelse(grepl("stevor-like", description), "stevor, pseudogene", description)) %>% -->
<!-- mutate(description = ifelse(grepl("exported protein family", description), "exported protein family", description)) %>% -->
<!-- mutate(description = ifelse(grepl("ribosomal RNA", description), "rRNA", description)) %>% -->
<!-- mutate(description = ifelse(grepl("serine/threonine protein kinase", description), "serine/threonine protein kinase, FIKK family", description)) %>% -->
<!-- mutate(description = ifelse(grepl("hypothetical protein", description), "hypothetical protein, conserved", description)) %>% -->
<!-- mutate(description = ifelse(grepl("conserved Plasmodium protein, unknown function", description), "hypothetical protein, conserved", description)) %>% -->
<!-- mutate(description = ifelse(grepl("Rifin/stevor family, putative", description), "stevor", description)) %>% -->
<!-- mutate(description = ifelse(grepl("stevor", description), "stevor", description)) %>% -->
<!-- mutate(description = ifelse(grepl("rifin", description), "rifin", description)) %>% -->
<!-- mutate(description = ifelse(grepl("erythrocyte membrane protein 1 (PfEMP1), pseudogene", description), "erythrocyte membrane protein 1 (PfEMP1)", description)) %>% -->
<!-- mutate(description = ifelse(grepl("erythrocyte membrane protein 1", description), "erythrocyte membrane protein 1 (PfEMP1)", description)) %>% -->
<!-- mutate(description = ifelse(grepl("probably protein", description), "unspecified product", description)) %>% -->
<!-- mutate(description = ifelse(grepl("RESA", description), "RESA", description)) %>% -->
<!-- mutate(description = ifelse(grepl("ring-infected erythrocyte surface antigen", description), "ring-infected erythrocyte surface antigen", description))%>% -->
<!-- mutate(description = ifelse(grepl("Duffy binding domain/Erythrocyte binding antigen175, putative", description), "erythrocyte binding like protein 1", description)) %>% -->
<!-- mutate(description = ifelse(grepl("erythrocyte binding like protein 1", description), "erythrocyte binding like protein 1", description))%>% -->
<!-- mutate(description = ifelse(grepl("unspecified product", description), "hypothetical protein, conserved", description))%>% -->
<!-- mutate(description = ifelse(grepl("probable protein, unknown function", description), "hypothetical protein, conserved", description)) %>% -->
<!-- mutate(description = ifelse(grepl("RESA", description), "ring-infected erythrocyte surface antigen", description)) -->
<!-- descriptionColorsNames = unique(c(allGenes$description)) -->
<!-- descriptionColors = scheme$hex(length(descriptionColorsNames)) -->
<!-- names(descriptionColors) = descriptionColorsNames -->
<!-- renamed_combined_sharedRegionsNotInTandems = readr::read_tsv("../../sharedBetween11_and_13/windows/renamed_combined_sharedRegionsNotInTandems.bed", col_names = F) -->
<!-- combined_11_13_sharedRegions_stepped = readr::read_tsv("../../sharedBetween11_and_13/windows/combined_11_13_sharedRegions_stepped.bed", col_names = F); -->
<!-- inbetweenConservedRegions = readr::read_tsv("../../sharedBetween11_and_13/windows/inbetweenConservedRegions.bed", col_names = F); -->
<!-- sharedRegionsNotInTandems_size150_step25 = readr::read_tsv("../../sharedBetween11_and_13/windows/sharedRegionsNotInTandems_size150_step25.bed", col_names = F); -->
<!-- bedsOfSharedRegions = readr::read_tsv("../../sharedBetween11_and_13/bedsOfSharedRegions/Pf3D7.bed", col_names = T); -->
<!-- inbetweenLargeTandems = readr::read_tsv("../../windows/inbetweenLargeTandems.bed", col_names = F); -->
<!-- finalWindows_inSharedRegion = readr::read_tsv("../windowAnalysis/windows/finalHRPII_HRPIII_windows.bed", col_names = F) %>% -->
<!-- left_join(outBed %>% -->
<!-- select(1:3) %>% -->
<!-- rename(X1 = `#chrom`, -->
<!-- sharedStart = start, -->
<!-- sharedEnd = end)) %>% -->
<!-- filter((X2 >= sharedStart & X2 < sharedEnd) | (X3 >= sharedStart & X3 <= sharedEnd)) -->
<!-- finalSubWindows_inSharedRegion = readr::read_tsv("../windowAnalysis/windows/finalHRPII_HRPIII_windows_combinedVarConservedRegions.bed", col_names = F) %>% -->
<!-- left_join(outBed %>% -->
<!-- select(1:3) %>% -->
<!-- rename(X1 = `#chrom`, -->
<!-- sharedStart = start, -->
<!-- sharedEnd = end)) %>% -->
<!-- filter((X2 >= sharedStart & X2 < sharedEnd) | (X3 >= sharedStart & X3 <= sharedEnd)) -->
<!-- allGenes_inSharedRegion = allGenes %>% -->
<!-- left_join(outBed %>% -->
<!-- select(1:3) %>% -->
<!-- rename(chrom = `#chrom`, -->
<!-- sharedStart = start, -->
<!-- sharedEnd = end)) %>% -->
<!-- filter((start >= sharedStart & start < sharedEnd) | (end >= sharedStart & end <= sharedEnd)) %>% -->
<!-- group_by(chrom, start, end, name, length) %>% -->
<!-- mutate(start = max(start, sharedStart), -->
<!-- end = min(end, sharedEnd)) -->
<!-- genomic_combined_filtered = readr::read_tsv("chroms11_13_regions_exact/sequences/all_trfOutput/filtered_genomic_combined.bed", col_names = F) -->
<!-- genomic_combined_filtered_3d7 = genomic_combined_filtered %>% -->
<!-- filter(X1 %in% c("Pf3D7_11_v3", "Pf3D7_13_v3")) -->
<!-- colnames(genomic_combined_filtered_3d7)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- colnames(finalWindows_inSharedRegion)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- colnames(finalSubWindows_inSharedRegion)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- colnames(allGenes_inSharedRegion)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- colnames(renamed_combined_sharedRegionsNotInTandems)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- colnames(combined_11_13_sharedRegions_stepped)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- colnames(inbetweenConservedRegions)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- colnames(sharedRegionsNotInTandems_size150_step25)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- colnames(bedsOfSharedRegions)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- colnames(inbetweenLargeTandems)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- inbetweenLargeTandems_inSharedRegion = inbetweenLargeTandems%>% -->
<!-- left_join(outBed %>% -->
<!-- select(1:3) %>% -->
<!-- rename( -->
<!-- sharedStart = start, -->
<!-- sharedEnd = end)) %>% -->
<!-- filter((start >= sharedStart & start < sharedEnd) | (end >= sharedStart & end <= sharedEnd)) %>% -->
<!-- group_by(`#chrom`, start, end, name, len) %>% -->
<!-- mutate(start = max(start, sharedStart), -->
<!-- end = min(end, sharedEnd)) %>% -->
<!-- ungroup() -->
<!-- # temp = readr::read_tsv("tunning/shared_11_13_region_trfOutput/onlymapped_nochr11Locs_windowsAcrossDuplicatedRegion_chr11_step20_size200_id90.bed", col_names = F) %>% -->
<!-- # group_by(X4) %>% -->
<!-- # count() %>% -->
<!-- # ungroup() %>% -->
<!-- # separate(X4, remove = F, convert = T, sep = "-", into = c("chrom", "start", "end", "strand")) %>% -->
<!-- # mutate("#chrom" = chrom) -->
<!-- # temp = readr::read_tsv("outFilt_id80.tab.txt", col_names = T) %>% -->
<!-- # mutate("#chrom" = chrom) -->
<!-- # -->
<!-- # temp2 = readr::read_tsv("outFilt_id90.tab.txt", col_names = T) %>% -->
<!-- # mutate("#chrom" = chrom) -->
<!-- temp = readr::read_tsv("filtered_id80_chr11_chr13_inDuplicatedArea_withGeneInfo.bed", col_names = F) -->
<!-- colnames(temp)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- regions_withSD01MultiInShared = readr::read_tsv("../windowAnalysis/regions_withSD01MultiInShared.bed") -->
<!-- new_regions_withSD01MultiInShared = readr::read_tsv("/Users/nick/projects/plasmodium/falciparum/PathWeaverAnalys_2020_12/chr11_chr13_inDuplicatedArea/PfSD01_chr11_chr13_inDuplicatedArea/final/hold.bed", col_names = F) -->
<!-- colnames(new_regions_withSD01MultiInShared)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- firstPassSuccess = readr::read_tsv("chrom11_13DuplicatedRegion_successfulFirstPass.bed", col_names = F) -->
<!-- colnames(firstPassSuccess)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- varWindowFromFirstPassSuccess = readr::read_tsv("finalTunned_duplicatedRegionChr11Chr13_combinedVarConservedRegions.bed", col_names = F) -->
<!-- colnames(varWindowFromFirstPassSuccess)[1:length(colnames(outBed))] = colnames(outBed) -->
<!-- ``` -->
<!-- ```{r} -->
<!-- #| fig-column: screen-inset-shaded -->
<!-- overlayPlot = ggplot() + -->
<!-- geom_vline(aes(xintercept = realPosition), -->
<!-- data = variableSitesInPacbioGenomes, -->
<!-- color = "red") + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = 0.5, ymax = 1), -->
<!-- data = outBed) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = 1, ymax = 2), -->
<!-- data = genomic_combined_filtered_3d7) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = 2, ymax = 3), fill = "maroon", -->
<!-- data = inbetweenLargeTandems_inSharedRegion) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = 3, ymax = 4), fill = "darkblue", -->
<!-- data = temp ) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = 3.75, ymax = 4), fill = "#00FCCF", -->
<!-- data = firstPassSuccess ) + -->
<!-- # geom_rect(aes(xmin = start, xmax = end, -->
<!-- # ymin = 4, ymax = 5), fill = "darkblue", -->
<!-- # data = temp2 ) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = 3, ymax = 3.5), fill = "limegreen", -->
<!-- data = regions_withSD01MultiInShared ) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = 3, ymax = 3.25), fill = "forestgreen", -->
<!-- data = new_regions_withSD01MultiInShared ) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = -1, ymax = -0.5), -->
<!-- data = finalWindows_inSharedRegion) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = -0.5, ymax = 0), fill = "orange", -->
<!-- data = finalSubWindows_inSharedRegion) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = 4, ymax = 5), fill = "orange", -->
<!-- data = finalSubWindows_inSharedRegion) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = 0, ymax = 1, -->
<!-- fill = description), -->
<!-- data = allGenes_inSharedRegion) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = -0.75, ymax = -0.25), fill = "blue", -->
<!-- data = combined_11_13_sharedRegions_stepped)+ -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = -0.75, ymax = -0.5), fill = "red", -->
<!-- data = renamed_combined_sharedRegionsNotInTandems)+ -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = -1, ymax = -2), fill = "purple", -->
<!-- data = inbetweenConservedRegions) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = -2.5, ymax = -2), fill = "lightblue", -->
<!-- data = bedsOfSharedRegions)+ -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = -3, ymax = -2.5), fill = "magenta", -->
<!-- data = sharedRegionsNotInTandems_size150_step25) + -->
<!-- geom_rect(aes(xmin = start, xmax = end, -->
<!-- ymin = 4, ymax = 4.25), fill = "#000000", -->
<!-- data = varWindowFromFirstPassSuccess )+ -->
<!-- scale_fill_manual(values = descriptionColors) + -->
<!-- facet_wrap(~`#chrom`, scales = "free", ncol = 1) + -->
<!-- sofonias_theme -->
<!-- print(overlayPlot) -->
<!-- ``` -->
<!-- ```{r} -->
<!-- #| column: screen-inset-shaded -->
<!-- ggplotly(overlayPlot) -->
<!-- ``` -->