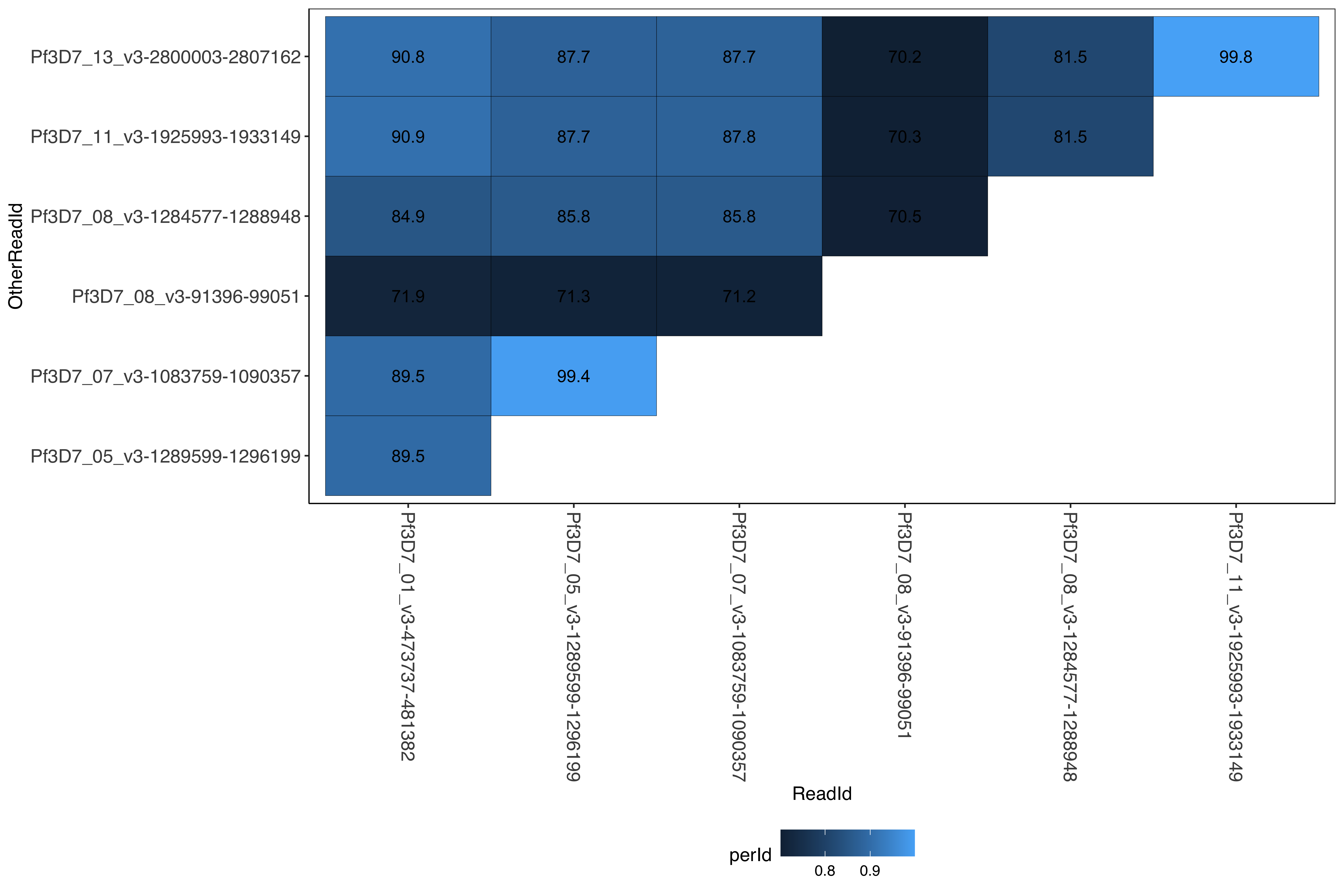
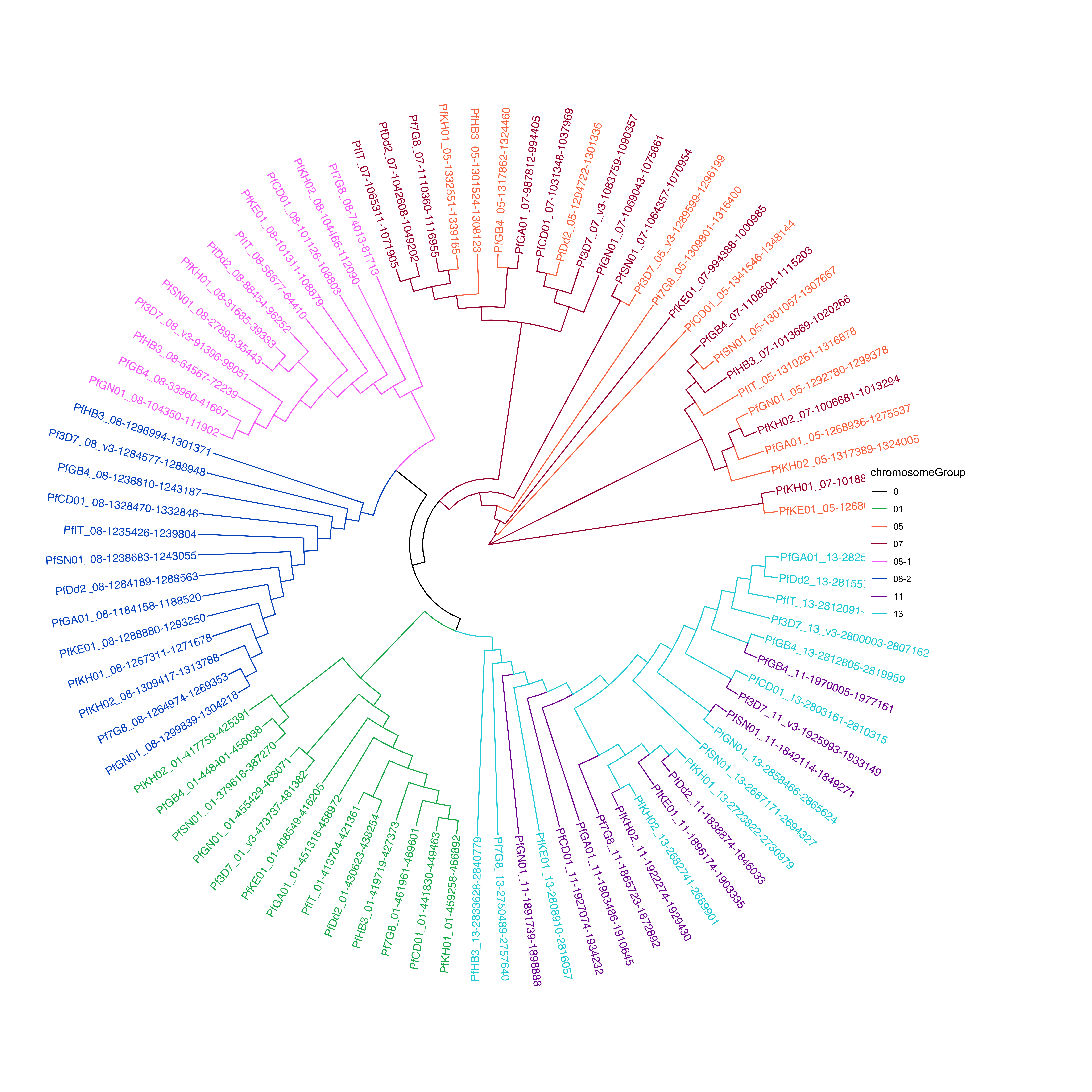
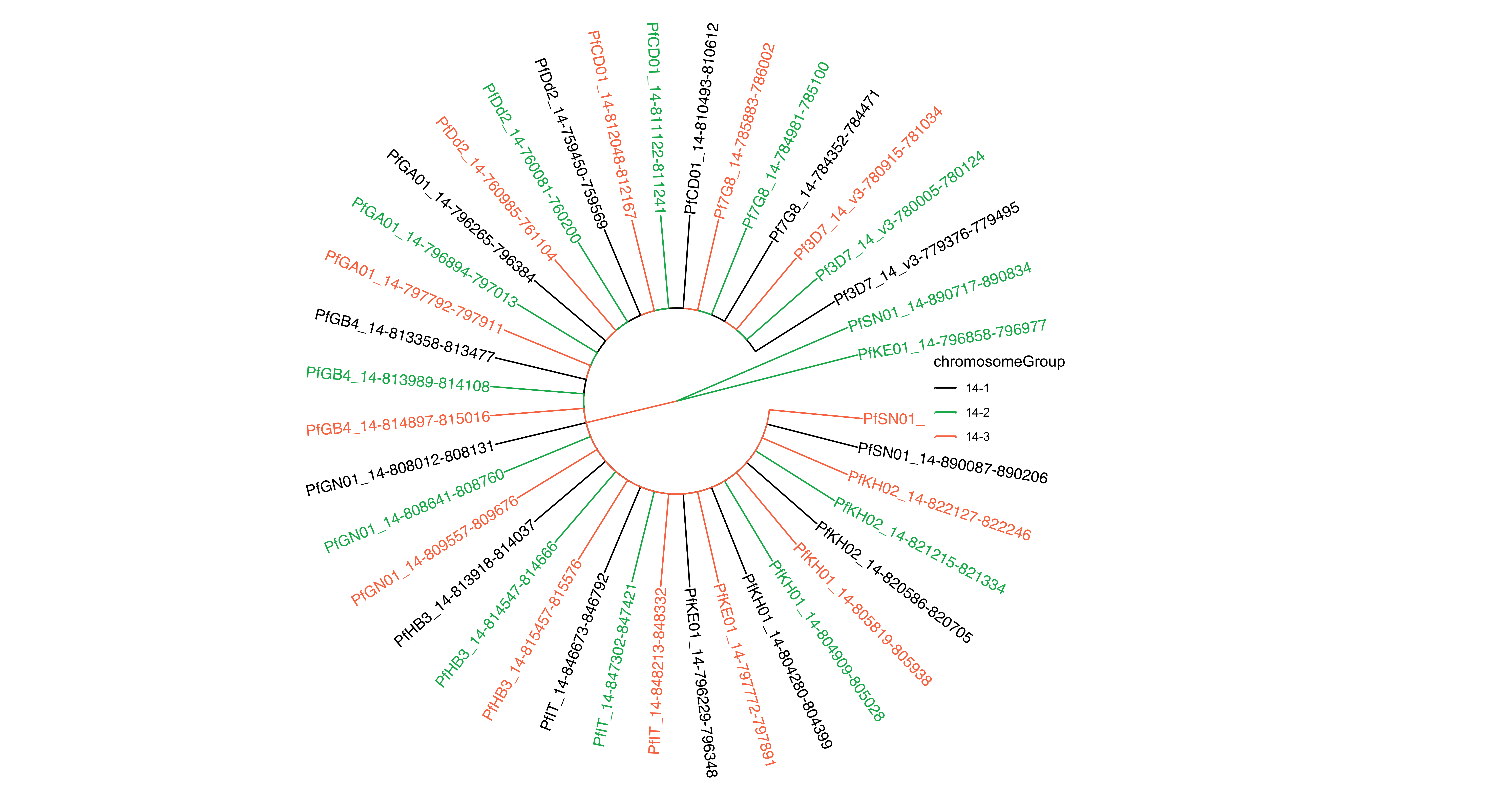
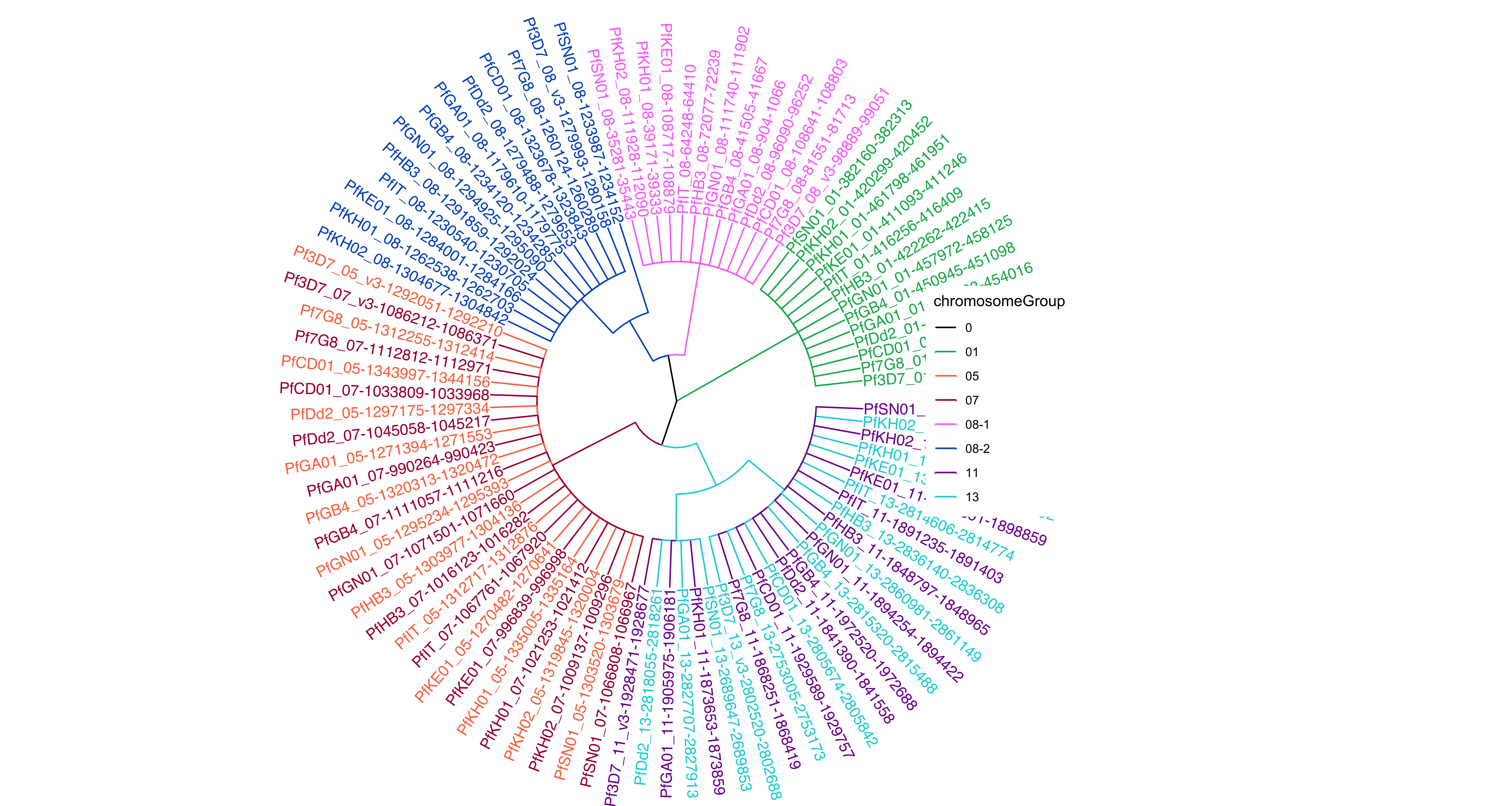
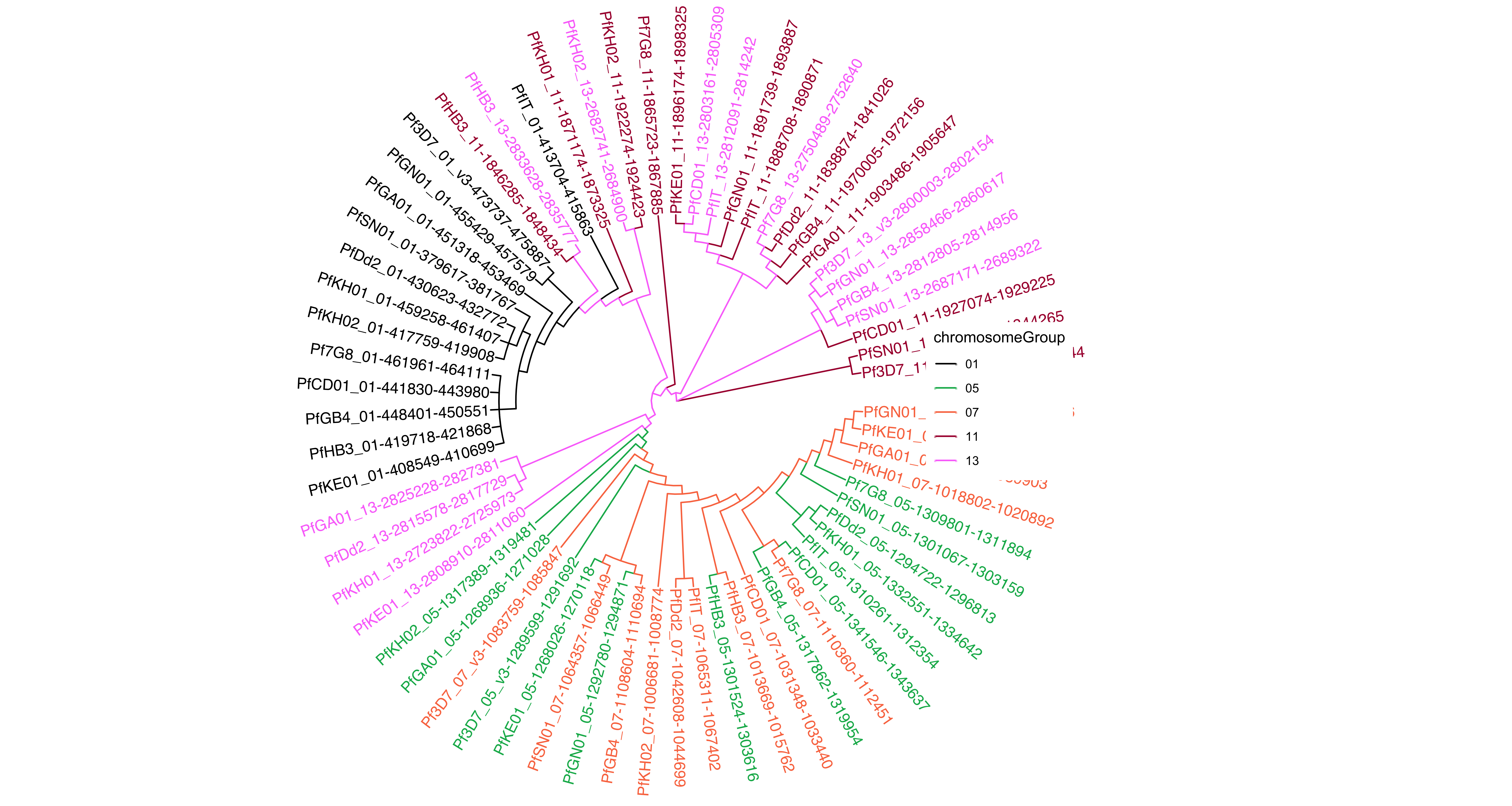
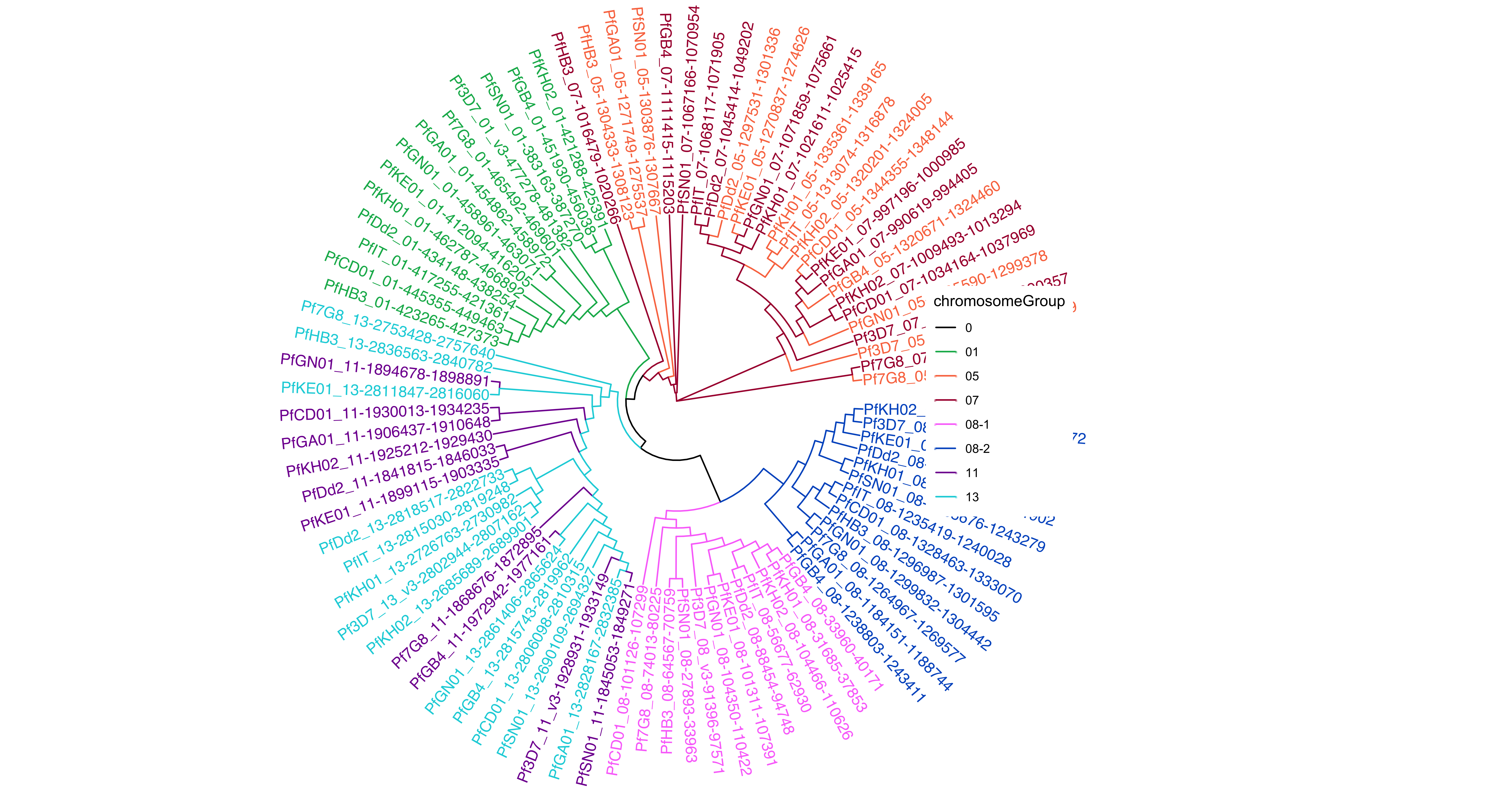
--- title: Info on 3D7 rRNA --- ```{r setup, echo=FALSE, message=FALSE} source("../../common.R") ``` ## Getting the larger organization sub unit ## Summary * two types of rRNA genes in P. falciparum encoded in 7 loci * S-type * expressed primarily while in the mosquito vector * Full 18S-5.8S-28S rRNA units * on chromosome 11 and 13 * A-type * expressed primarily while in the human host * Full 18S-5.8S-28S rRNA units * on chromosome 5 and 7 * Mix-type * On chromosome 1 * 18S-5.8S rRNAs identical to S-type but a 28S rRNA unit that is 65% identity to the A-type and 75% identity to the S-type * Partial * chromosome 8 two unusual rRNA gene units that contain 5.8S and 28S rRNA units but no 18s rRNAs * unknown if these are functional * 5S rRNA * three identical tandemly arrayed units on chromosome 14 ```{bash, eval = F} elucidator gffDescriptionsCount --gff /tank/data/genomes/plasmodium/genomes/pf/info/gff/Pf3D7.gff | egrep "ribosomal" | egrep "RNA" | tail -4 | cut -f1 > rRNA_descriptionsUpdated.txt elucidator gffToBedByDescription --gff /tank/data/genomes/plasmodium/genomes/pf/info/gff/Pf3D7.gff --description rRNA_descriptionsUpdated.txt --features gene --out updatedrRNA_locs elucidator getFastaWithBed --bed updatedrRNA_locs.bed --twoBit /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.2bit --out updatedrRNA_locs.fasta --overWrite bedtools merge -d 2000 -i updatedrRNA_locs.bed | elucidator bed3ToBed6 --bed STDIN | elucidator filterBedRecordsByLength --minLen 2000 --bed STDIN --overWrite | elucidator reorientBedToIntersectingGeneInGff --bed STDIN --gff /tank/data/genomes/plasmodium/genomes/pf/info/gff/Pf3D7.gff --out merged_updatedrRNA_locs.bed --overWrite # against reichnowi elucidator extractRefSeqsFromGenomes --bed merged_updatedrRNA_locs.bed --genomeDir /tank/data/genomes/plasmodium/genomes/plasmodiumRefGenomes/genomes/ --gffDir /tank/data/genomes/plasmodium/genomes/plasmodiumRefGenomes/info/gff/ --primaryGenome Pf3D7 --outputDir pr_extract_rRNA_regions --numThreads 15 --overWriteDirs --selectedGenomes PRG01,Pf3D7 # references elucidator extractRefSeqsFromGenomes --bed merged_updatedrRNA_locs.bed --genomeDir /tank/data/genomes/plasmodium/genomes/plasmodiumRefGenomes/genomes/ --gffDir /tank/data/genomes/plasmodium/genomes/plasmodiumRefGenomes/info/gff/ --primaryGenome Pf3D7 --outputDir primaryReference_extract_rRNA_regions --numThreads 15 --overWriteDirs elucidator extractRefSeqsFromGenomes --bed merged_updatedrRNA_locs.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pf/genomes/ --gffDir /tank/data/genomes/plasmodium/genomes/pf/info/gff/ --primaryGenome Pf3D7 --outputDir extract_rRNA_regions --numThreads 15 --overWriteDirs cd extract_rRNA_regions cat */beds/*.bed | elucidator bedCoordSort --bed STDIN | cut -f1-6 | bedtools merge | egrep -v _00_ | elucidator bed3ToBed6 --bed STDIN | elucidator reorientBedToIntersectingGeneInGff --bed STDIN --gff /tank/data/genomes/combinedGenomes/allPf/info/gff/allPf.gff --overWrite --out allrRNA_locs.bed elucidator getFastaWithBed --bed allrRNA_locs.bed --twoBit /tank/data/genomes/combinedGenomes/allPfRaw/genomes/allPfRaw.2bit --out allrRNA_locs.fasta --overWrite muscle -in allrRNA_locs.fasta -out aln_allrRNA_locs.fasta FastTree -nt aln_allrRNA_locs.fasta > aln_allrRNA_locs.nwk # all by all compare for percent identity elucidator compareAllByAll --fasta allrRNA_locs.fasta --out allByAll.tab.txt --numThreads 15 --overWrite --verbose --alnInfoDir alnCache ``` # Genomic locations of rRNA loci/subunits ```{r} = readr:: read_tsv ("allGenomicPfrRNA.bed" , col_names = F):: datatable (regions,extensions = 'Buttons' , options = list (dom = 'Bfrtip' ,buttons = c ('csv' )``` # All by all percent identiy between the different rRNA loci ```{r} = readr:: read_tsv ("extract_rRNA_regions/allByAll.tab.txt" )= allByAll %>% filter (grepl ("Pf3D7" , ReadId))%>% filter (grepl ("Pf3D7" , OtherReadId)) %>% mutate (OtherReadId = factor (OtherReadId, levels = naturalsort:: naturalsort (unique (c (.$ ReadId, .$ OtherReadId))))) %>% mutate (ReadId = factor (ReadId, levels = naturalsort:: naturalsort (unique (c (.$ ReadId, .$ OtherReadId))))) ggplot (allByAll_3d7) + geom_tile (aes (x = ReadId, y = OtherReadId, fill = perId), color = "black" ) + geom_text (aes (x = ReadId, y = OtherReadId, label = round (perId * 100 , 1 ))) + ``` # Full unit ```{r} #| fig-column: screen #| fig-height: 15 #| fig-width: 15 library (ggtree)= ape:: read.tree ("extract_rRNA_regions/aln_allrRNA_locs.nwk" )= tibble (seq = rRna_tree$ tip.label) %>% mutate (chromNum = gsub ("-.*" , "" , seq)) %>% mutate (chromNum = gsub ("_v3" , "" , chromNum)) %>% mutate (chromNum = gsub (".*_" , "" , chromNum)) %>% mutate (genome = gsub ("_.*" , "" , seq)) %>% separate (seq, into = c ("chrom" , "start" , "end" ), sep = "-" , convert = T, remove = F) %>% arrange (genome, chrom, start) %>% group_by (genome, chromNum) %>% mutate (chromID = row_number (), total = n ()) %>% mutate (chromLabel = ifelse (total > 1 , paste0 (chromNum, "-" , chromID), chromNum ) ) %>% mutate (chromLabel = ifelse ("PfGA01_08-1184158-1188520" == seq, "08-2" , chromLabel))= list ()for (currentChrom in unique (rRna_tree_labelDf$ chromLabel)){= rRna_tree_labelDf %>% filter (chromLabel == currentChrom)= rRna_tree_labelDf_chrom$ seq= groupOTU (rRna_tree, chroms)print (ggtree (rRna_tree, layout = 'circular' , branch.length= 'none' , aes (color = group)) + geom_tiplab () + scale_color_manual ("chromosomeGroup" , values = c ("black" , "#0AB45A" , "#FA7850" , "#AA0A3C" , "#FA78FA" , "#005AC8" , "#8214A0" , "#14D2DC" ), guide = guide_legend (override.aes= list (shape= 1 , size = 1 ))) + hexpand (0.5 ) + theme (plot.margin= margin (120 , 120 , 120 , 120 )))pdf ("rRNA_tree.pdf" , width = 11 , height = 11 , useDingbats = F)print (ggtree (rRna_tree, layout = 'circular' , branch.length= 'none' , aes (color = group)) + geom_tiplab () + scale_color_manual ("chromosomeGroup" , values = c ("black" , "#0AB45A" , "#FA7850" , "#AA0A3C" , "#FA78FA" , "#005AC8" , "#8214A0" , "#14D2DC" ), guide = guide_legend (override.aes= list (shape= 1 , size = 1 ))) + hexpand (0.5 ) + theme (plot.margin= margin (120 , 120 , 120 , 120 )))# print(ggtree(rRna_tree, layout = 'circular', aes(color = group)) + # geom_tiplab() + # scale_color_manual("chromosomeGroup", values = c("black", "#0AB45A", "#FA7850", "#AA0A3C", "#FA78FA", "#005AC8", "#8214A0", "#14D2DC"), # guide = guide_legend(override.aes=list(shape=1, size =1))) + hexpand(0.5) + theme(plot.margin=margin(120, 120, 120, 120))) dev.off ()``` # By subunit ## 5s ```{bash, eval = F} elucidator gffToBedByDescription --gff /tank/data/genomes/plasmodium/genomes/pf/info/gff/Pf3D7.gff --description 5s_rRNA_descriptionsUpdated.txt --features gene --out 5s_rRNA_locs.bed elucidator getFastaWithBed --bed 5s_rRNA_locs.bed --twoBit /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.2bit --out 5s_rRNA_locs.fasta --overWrite elucidator extractRefSeqsFromGenomes --bed 5s_rRNA_locs.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pf/genomes/ --gffDir /tank/data/genomes/plasmodium/genomes/pf/info/gff/ --primaryGenome Pf3D7 --outputDir 5s_rRNA_locs_regions --numThreads 15 --overWriteDirs cd 5s_rRNA_locs_regions cat */beds/*.bed | elucidator bedCoordSort --bed STDIN | cut -f1-6 | bedtools merge | egrep -v _00_ | elucidator bed3ToBed6 --bed STDIN --overWrite --out allrRNA_locs.bed elucidator getFastaWithBed --bed allrRNA_locs.bed --twoBit /tank/data/genomes/combinedGenomes/allPfRaw/genomes/allPfRaw.2bit --out allrRNA_locs.fasta --overWrite muscle -in allrRNA_locs.fasta -out aln_allrRNA_locs.fasta FastTree -nt aln_allrRNA_locs.fasta > aln_allrRNA_locs.nwk ``` ```{r} #| fig-column: screen #| #| fig-height: 15 #| fig-width: 15 library (ggtree)= ape:: read.tree ("5s_rRNA_locs_regions/aln_allrRNA_locs.nwk" )= tibble (seq = rRna_tree$ tip.label) %>% mutate (chromNum = gsub ("-.*" , "" , seq)) %>% mutate (chromNum = gsub ("_v3" , "" , chromNum)) %>% mutate (chromNum = gsub (".*_" , "" , chromNum)) %>% mutate (genome = gsub ("_.*" , "" , seq)) %>% separate (seq, into = c ("chrom" , "start" , "end" ), sep = "-" , convert = T, remove = F) %>% arrange (genome, chrom, start) %>% group_by (genome, chromNum) %>% mutate (chromID = row_number (), total = n ()) %>% mutate (chromLabel = ifelse (total > 1 , paste0 (chromNum, "-" , chromID), chromNum ) ) = list ()for (currentChrom in unique (rRna_tree_labelDf$ chromLabel)){= rRna_tree_labelDf %>% filter (chromLabel == currentChrom)= rRna_tree_labelDf_chrom$ seq= groupOTU (rRna_tree, chroms)= ggtree (rRna_tree, layout = 'circular' , branch.length= 'none' , aes (color = group)) + geom_tiplab () + scale_color_manual ("chromosomeGroup" , values = c ("black" , "#0AB45A" , "#FA7850" , "#AA0A3C" , "#FA78FA" , "#005AC8" , "#8214A0" , "#14D2DC" ), guide = guide_legend (override.aes= list (shape= 1 , size = 1 ))) + hexpand (0.5 ) + theme (plot.margin= margin (120 , 120 , 120 , 120 ))print (rRNA_tree_plot_5s)pdf ("5s_rRNA_tree.pdf" , width = 11 , height = 11 , useDingbats = F)print (rRNA_tree_plot_5s)dev.off ()``` ## 5.8s ```{bash, eval = F} elucidator gffToBedByDescription --gff /tank/data/genomes/plasmodium/genomes/pf/info/gff/Pf3D7.gff --description 5.8s_rRNA_descriptionsUpdated.txt --features gene --out 5.8s_rRNA_locs.bed elucidator getFastaWithBed --bed 5.8s_rRNA_locs.bed --twoBit /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.2bit --out 5.8s_rRNA_locs.fasta --overWrite elucidator extractRefSeqsFromGenomes --bed 5.8s_rRNA_locs.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pf/genomes/ --gffDir /tank/data/genomes/plasmodium/genomes/pf/info/gff/ --primaryGenome Pf3D7 --outputDir 5.8s_rRNA_locs_regions --numThreads 15 --overWriteDirs cd 5.8s_rRNA_locs_regions cat */beds/*.bed | elucidator bedCoordSort --bed STDIN | cut -f1-6 | bedtools merge | egrep -v _00_ | elucidator bed3ToBed6 --bed STDIN --overWrite --out allrRNA_locs.bed elucidator getFastaWithBed --bed allrRNA_locs.bed --twoBit /tank/data/genomes/combinedGenomes/allPfRaw/genomes/allPfRaw.2bit --out allrRNA_locs.fasta --overWrite muscle -in allrRNA_locs.fasta -out aln_allrRNA_locs.fasta FastTree -nt aln_allrRNA_locs.fasta > aln_allrRNA_locs.nwk ``` ```{r} #| fig-column: screen #| #| fig-height: 15 #| fig-width: 15 library (ggtree)= ape:: read.tree ("5.8s_rRNA_locs_regions/aln_allrRNA_locs.nwk" )= tibble (seq = rRna_tree$ tip.label) %>% mutate (chromNum = gsub ("-.*" , "" , seq)) %>% mutate (chromNum = gsub ("_v3" , "" , chromNum)) %>% mutate (chromNum = gsub (".*_" , "" , chromNum)) %>% mutate (genome = gsub ("_.*" , "" , seq)) %>% separate (seq, into = c ("chrom" , "start" , "end" ), sep = "-" , convert = T, remove = F) %>% arrange (genome, chrom, start) %>% group_by (genome, chromNum) %>% mutate (chromID = row_number (), total = n ()) %>% mutate (chromLabel = ifelse (total > 1 , paste0 (chromNum, "-" , chromID), chromNum ) ) = list ()for (currentChrom in unique (rRna_tree_labelDf$ chromLabel)){= rRna_tree_labelDf %>% filter (chromLabel == currentChrom)= rRna_tree_labelDf_chrom$ seq= groupOTU (rRna_tree, chroms).8 s = ggtree (rRna_tree, layout = 'circular' , branch.length= 'none' , aes (color = group)) + geom_tiplab () + scale_color_manual ("chromosomeGroup" , values = c ("black" , "#0AB45A" , "#FA7850" , "#AA0A3C" , "#FA78FA" , "#005AC8" , "#8214A0" , "#14D2DC" ), guide = guide_legend (override.aes= list (shape= 1 , size = 1 ))) + hexpand (0.5 ) + theme (plot.margin= margin (120 , 120 , 120 , 120 ))print (rRNA_tree_plot_5.8 s)pdf ("5.8s_rRNA_tree.pdf" , width = 11 , height = 11 , useDingbats = F)print (rRNA_tree_plot_5.8 s)dev.off ()``` ## 18s ```{bash, eval = F} elucidator gffToBedByDescription --gff /tank/data/genomes/plasmodium/genomes/pf/info/gff/Pf3D7.gff --description 18s_rRNA_descriptionsUpdated.txt --features gene --out 18s_rRNA_locs.bed elucidator getFastaWithBed --bed 18s_rRNA_locs.bed --twoBit /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.2bit --out 18s_rRNA_locs.fasta --overWrite elucidator extractRefSeqsFromGenomes --bed 18s_rRNA_locs.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pf/genomes/ --gffDir /tank/data/genomes/plasmodium/genomes/pf/info/gff/ --primaryGenome Pf3D7 --outputDir 18s_rRNA_locs_regions --numThreads 15 --overWriteDirs cd 18s_rRNA_locs_regions cat */beds/*.bed | elucidator bedCoordSort --bed STDIN | cut -f1-6 | bedtools merge | egrep -v _00_ | elucidator bed3ToBed6 --bed STDIN --overWrite --out allrRNA_locs.bed elucidator getFastaWithBed --bed allrRNA_locs.bed --twoBit /tank/data/genomes/combinedGenomes/allPfRaw/genomes/allPfRaw.2bit --out allrRNA_locs.fasta --overWrite muscle -in allrRNA_locs.fasta -out aln_allrRNA_locs.fasta FastTree -nt aln_allrRNA_locs.fasta > aln_allrRNA_locs.nwk ``` ```{r} #| fig-column: screen #| #| fig-height: 15 #| fig-width: 15 library (ggtree)= ape:: read.tree ("18s_rRNA_locs_regions/aln_allrRNA_locs.nwk" )= tibble (seq = rRna_tree$ tip.label) %>% mutate (chromNum = gsub ("-.*" , "" , seq)) %>% mutate (chromNum = gsub ("_v3" , "" , chromNum)) %>% mutate (chromNum = gsub (".*_" , "" , chromNum)) %>% mutate (genome = gsub ("_.*" , "" , seq)) %>% separate (seq, into = c ("chrom" , "start" , "end" ), sep = "-" , convert = T, remove = F) %>% arrange (genome, chrom, start) %>% group_by (genome, chromNum) %>% mutate (chromID = row_number (), total = n ()) %>% mutate (chromLabel = ifelse (total > 1 , paste0 (chromNum, "-" , chromID), chromNum ) ) = list ()for (currentChrom in unique (rRna_tree_labelDf$ chromLabel)){= rRna_tree_labelDf %>% filter (chromLabel == currentChrom)= rRna_tree_labelDf_chrom$ seq= groupOTU (rRna_tree, chroms)= ggtree (rRna_tree, layout = 'circular' , branch.length= 'none' , aes (color = group)) + geom_tiplab () + scale_color_manual ("chromosomeGroup" , values = c ("black" , "#0AB45A" , "#FA7850" , "#AA0A3C" , "#FA78FA" , "#005AC8" , "#8214A0" , "#14D2DC" ), guide = guide_legend (override.aes= list (shape= 1 , size = 1 ))) + hexpand (0.5 ) + theme (plot.margin= margin (120 , 120 , 120 , 120 )) print (rRNA_tree_plot_18s )pdf ("18s_rRNA_tree.pdf" , width = 11 , height = 11 , useDingbats = F)print (rRNA_tree_plot_18s+ theme (plot.margin= margin (120 , 120 , 120 , 120 )))dev.off ()``` ### With branch lengths ```{r} #| fig-column: screen #| #| fig-height: 15 #| fig-width: 15 = ggtree (rRna_tree, layout = 'circular' , aes (color = group)) + geom_tiplab () + scale_color_manual ("chromosomeGroup" , values = c ("black" , "#0AB45A" , "#FA7850" , "#AA0A3C" , "#FA78FA" , "#005AC8" , "#8214A0" , "#14D2DC" ), guide = guide_legend (override.aes= list (shape= 1 , size = 1 ))) + hexpand (0.5 ) + theme (plot.margin= margin (120 , 120 , 120 , 120 ))print (rRNA_tree_plot_18s_brLen)pdf ("18s_rRNA_tree_brLen.pdf" , width = 11 , height = 11 , useDingbats = F)print (rRNA_tree_plot_18s_brLen)dev.off ()``` ## 28s ```{bash, eval = F} elucidator gffToBedByDescription --gff /tank/data/genomes/plasmodium/genomes/pf/info/gff/Pf3D7.gff --description 28s_rRNA_descriptionsUpdated.txt --features gene --out 28s_rRNA_locs.bed elucidator getFastaWithBed --bed 28s_rRNA_locs.bed --twoBit /tank/data/genomes/plasmodium/genomes/pf/genomes/Pf3D7.2bit --out 28s_rRNA_locs.fasta --overWrite elucidator extractRefSeqsFromGenomes --bed 28s_rRNA_locs.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pf/genomes/ --gffDir /tank/data/genomes/plasmodium/genomes/pf/info/gff/ --primaryGenome Pf3D7 --outputDir 28s_rRNA_locs_regions --numThreads 15 --overWriteDirs cd 28s_rRNA_locs_regions cat */beds/*.bed | elucidator bedCoordSort --bed STDIN | cut -f1-6 | bedtools merge | egrep -v _00_ | elucidator bed3ToBed6 --bed STDIN --overWrite --out allrRNA_locs.bed elucidator getFastaWithBed --bed allrRNA_locs.bed --twoBit /tank/data/genomes/combinedGenomes/allPfRaw/genomes/allPfRaw.2bit --out allrRNA_locs.fasta --overWrite muscle -in allrRNA_locs.fasta -out aln_allrRNA_locs.fasta FastTree -nt aln_allrRNA_locs.fasta > aln_allrRNA_locs.nwk ``` ```{r} #| fig-column: screen #| #| fig-height: 15 #| fig-width: 15 library (ggtree)= ape:: read.tree ("28s_rRNA_locs_regions/aln_allrRNA_locs.nwk" )= tibble (seq = rRna_tree$ tip.label) %>% mutate (chromNum = gsub ("-.*" , "" , seq)) %>% mutate (chromNum = gsub ("_v3" , "" , chromNum)) %>% mutate (chromNum = gsub (".*_" , "" , chromNum)) %>% mutate (genome = gsub ("_.*" , "" , seq)) %>% separate (seq, into = c ("chrom" , "start" , "end" ), sep = "-" , convert = T, remove = F) %>% arrange (genome, chrom, start) %>% group_by (genome, chromNum) %>% mutate (chromID = row_number (), total = n ()) %>% mutate (chromLabel = ifelse (total > 1 , paste0 (chromNum, "-" , chromID), chromNum ) ) %>% mutate (chromLabel = ifelse ("PfGA01_08-1184151-1188744" == seq, "08-2" , chromLabel))= list ()for (currentChrom in unique (rRna_tree_labelDf$ chromLabel)){= rRna_tree_labelDf %>% filter (chromLabel == currentChrom)= rRna_tree_labelDf_chrom$ seq= groupOTU (rRna_tree, chroms)= ggtree (rRna_tree, layout = 'circular' , branch.length= 'none' , aes (color = group)) + geom_tiplab () + scale_color_manual ("chromosomeGroup" , values = c ("black" , "#0AB45A" , "#FA7850" , "#AA0A3C" , "#FA78FA" , "#005AC8" , "#8214A0" , "#14D2DC" ), guide = guide_legend (override.aes= list (shape= 1 , size = 1 ))) + hexpand (0.5 ) + theme (plot.margin= margin (120 , 120 , 120 , 120 ))print (rRNA_tree_plot_28s)pdf ("28s_rRNA_tree.pdf" , width = 11 , height = 11 , useDingbats = F)print (rRNA_tree_plot_28s)dev.off ()``` ### With branch lengths ```{r} #| fig-column: screen #| #| fig-height: 15 #| fig-width: 15 = ggtree (rRna_tree, layout = 'circular' , aes (color = group)) + geom_tiplab () + scale_color_manual ("chromosomeGroup" , values = c ("black" , "#0AB45A" , "#FA7850" , "#AA0A3C" , "#FA78FA" , "#005AC8" , "#8214A0" , "#14D2DC" ), guide = guide_legend (override.aes= list (shape= 1 , size = 1 ))) + hexpand (0.5 ) + theme (plot.margin= margin (120 , 120 , 120 , 120 ))print (rRNA_tree_plot_28s_brLen)pdf ("28s_rRNA_tree_brLen.pdf" , width = 11 , height = 11 , useDingbats = F)print (rRNA_tree_plot_28s_brLen)dev.off ()```