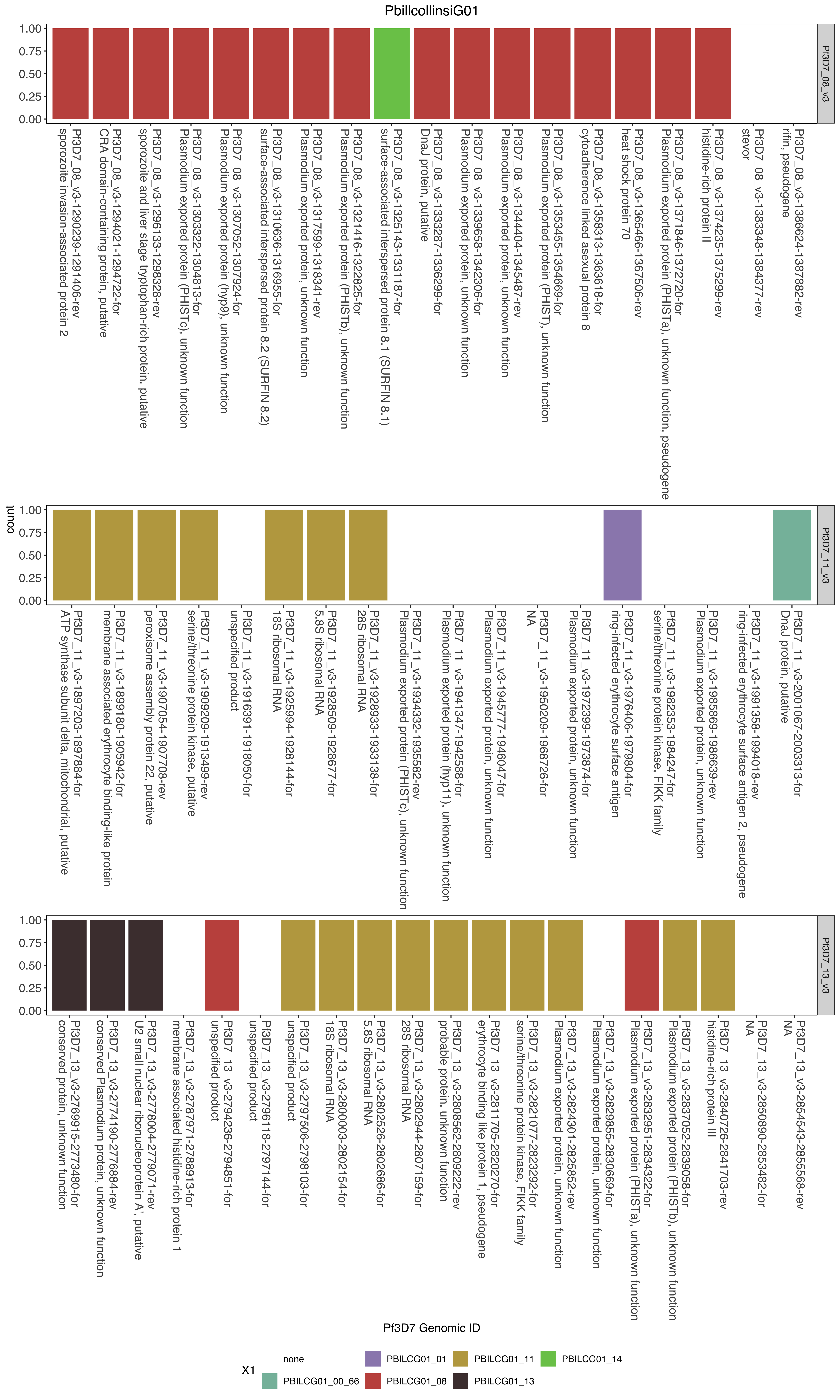
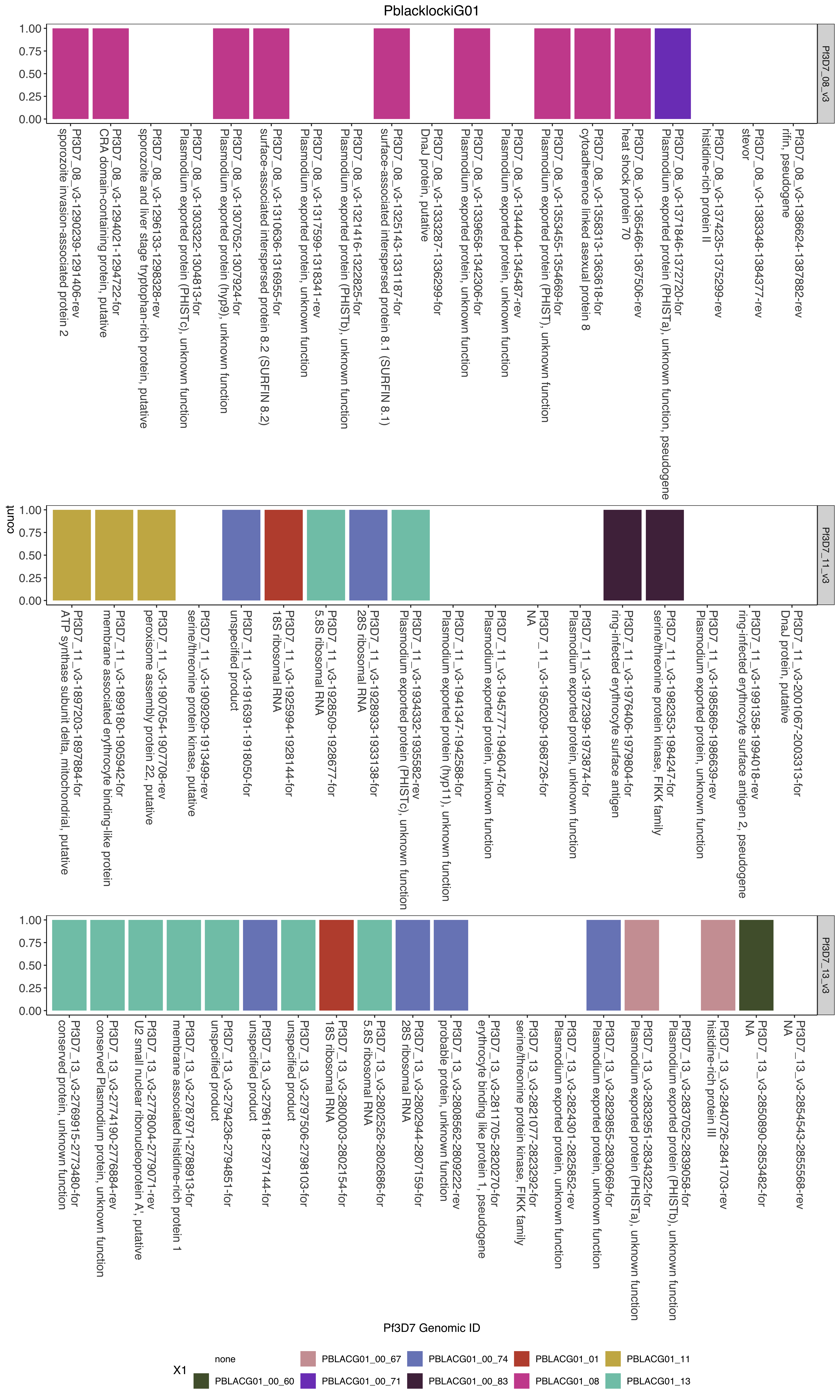
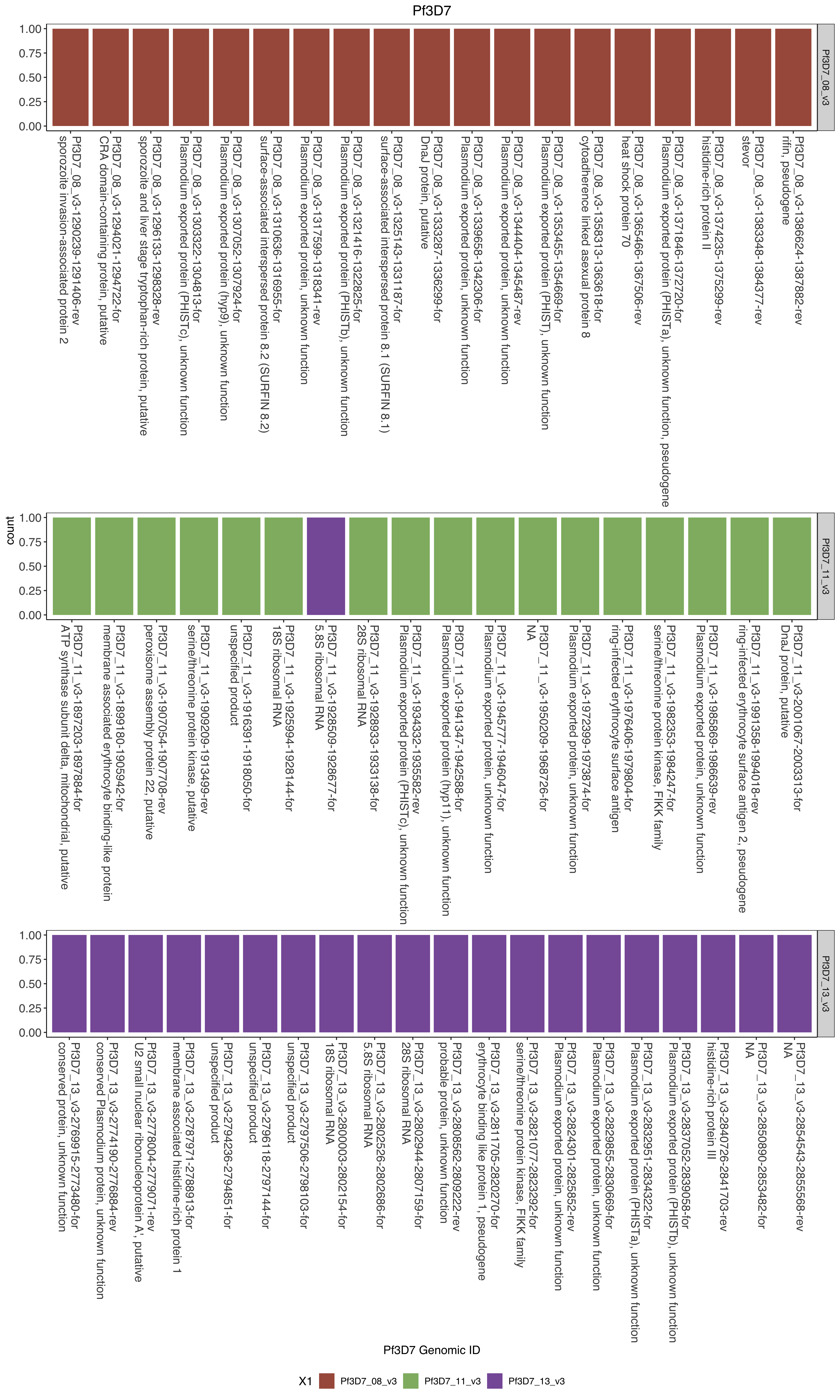
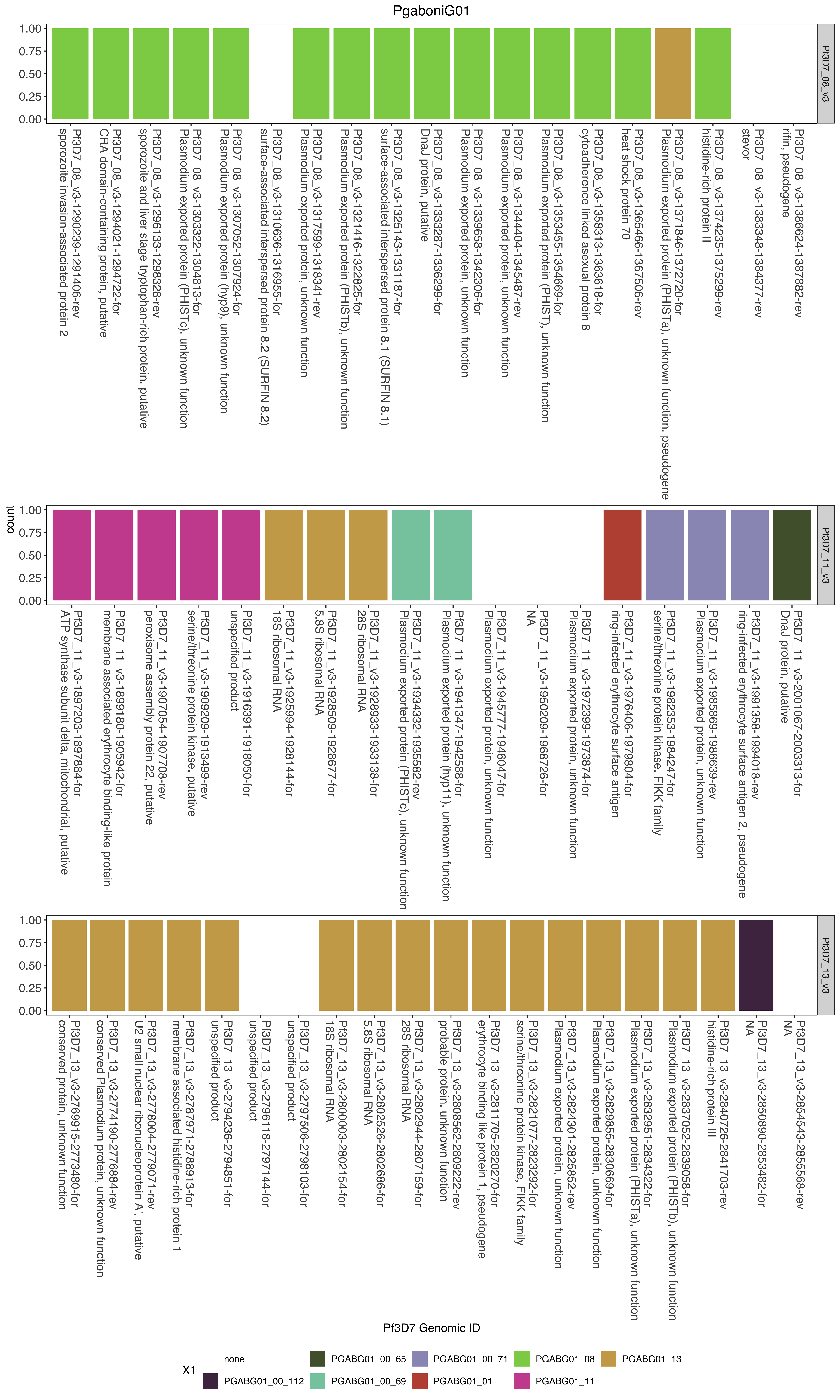
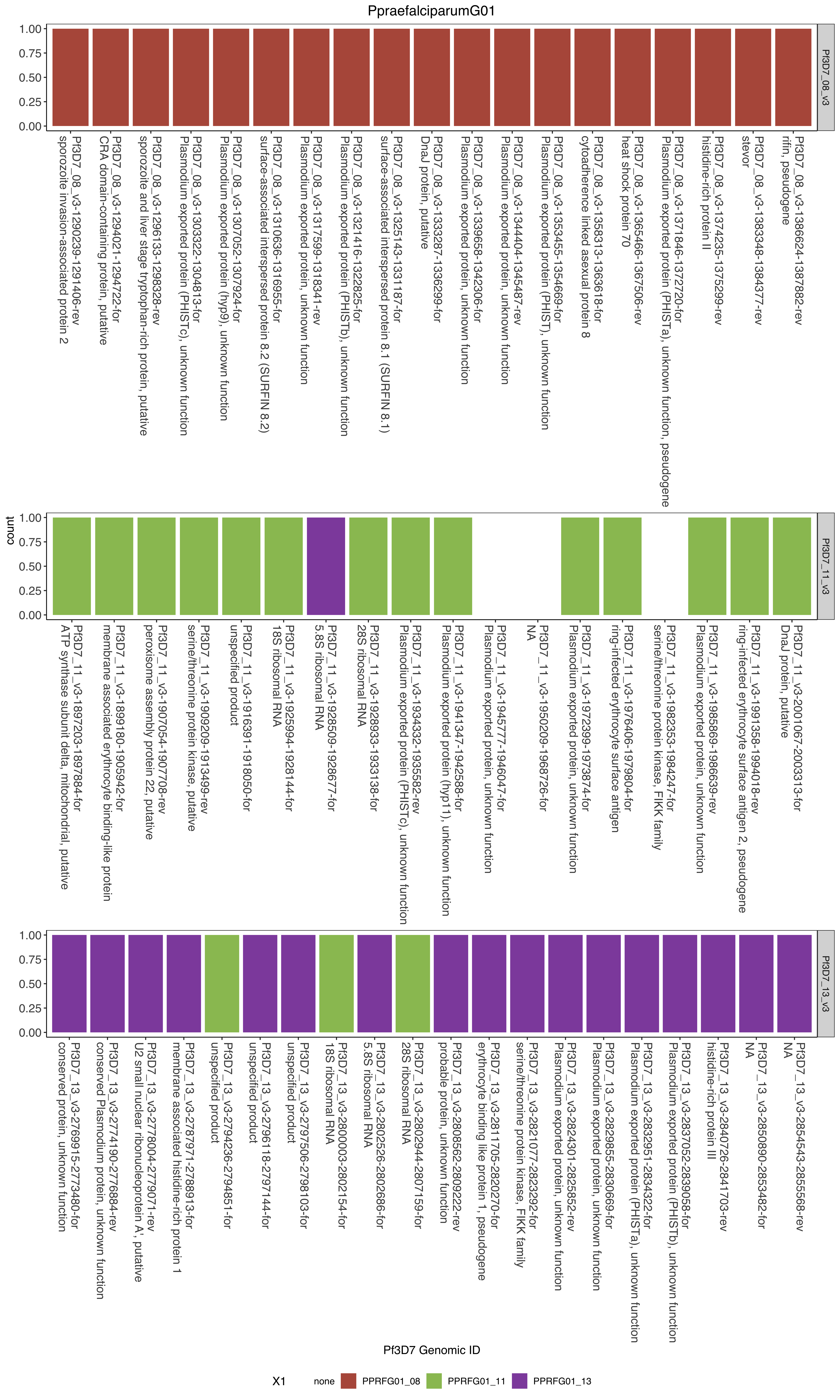
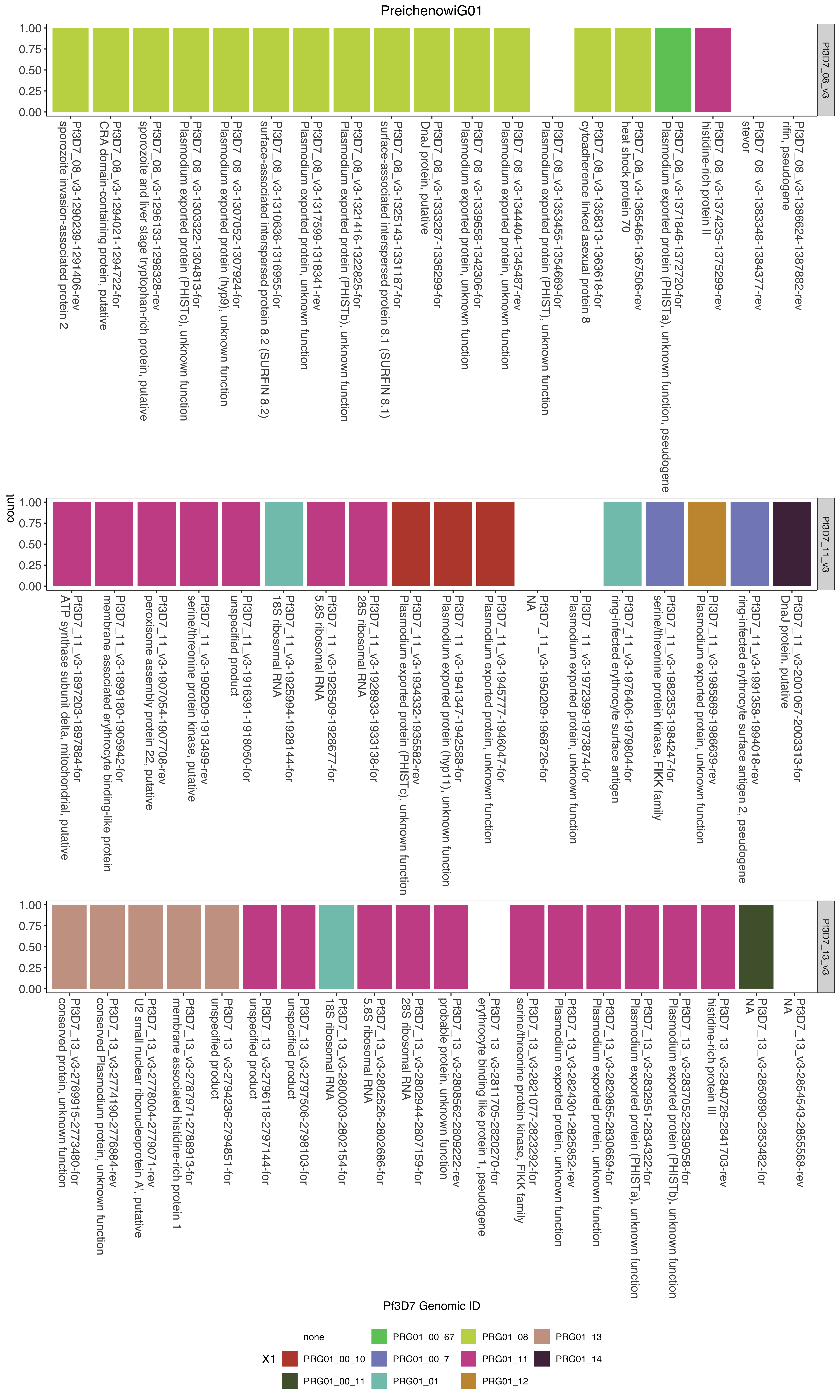
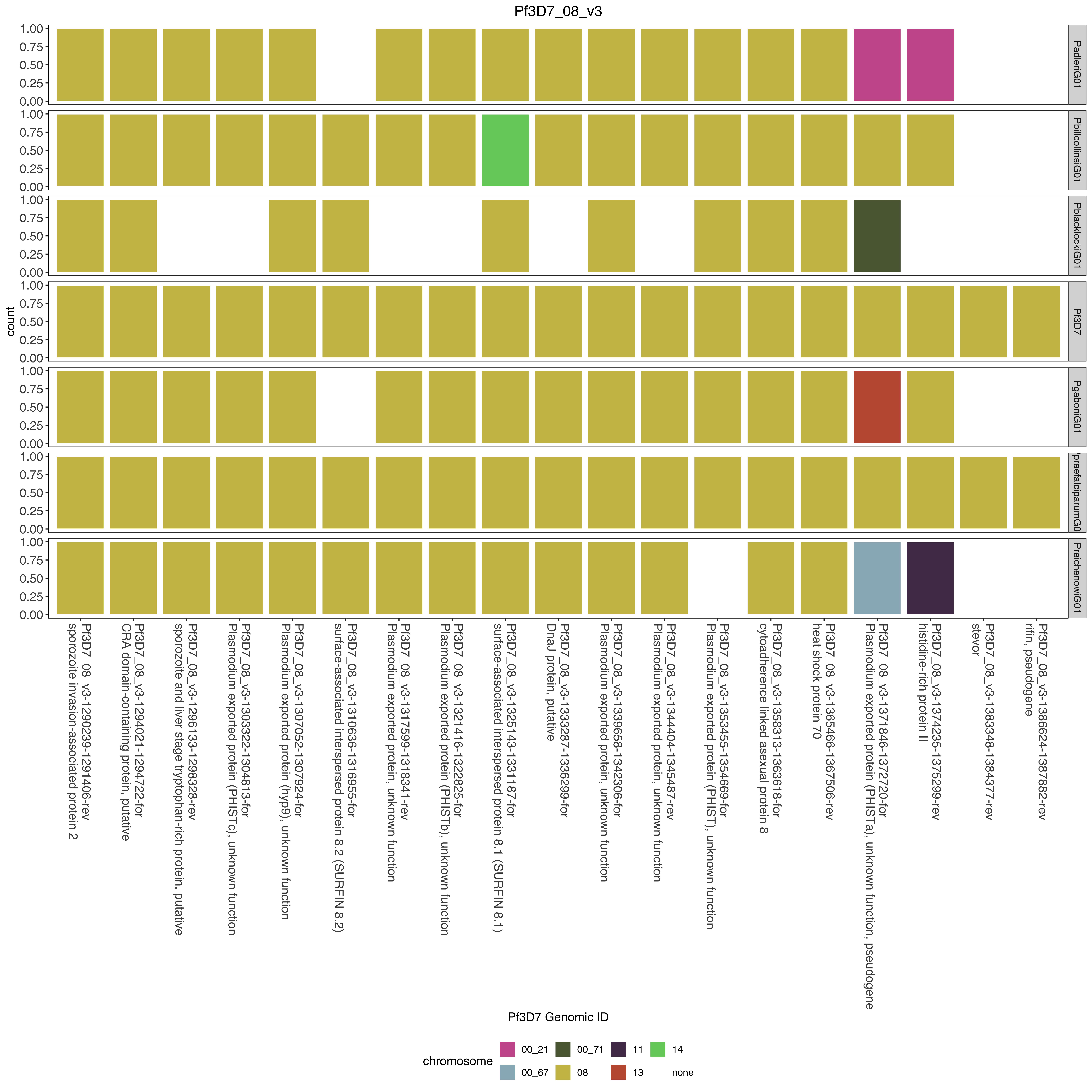
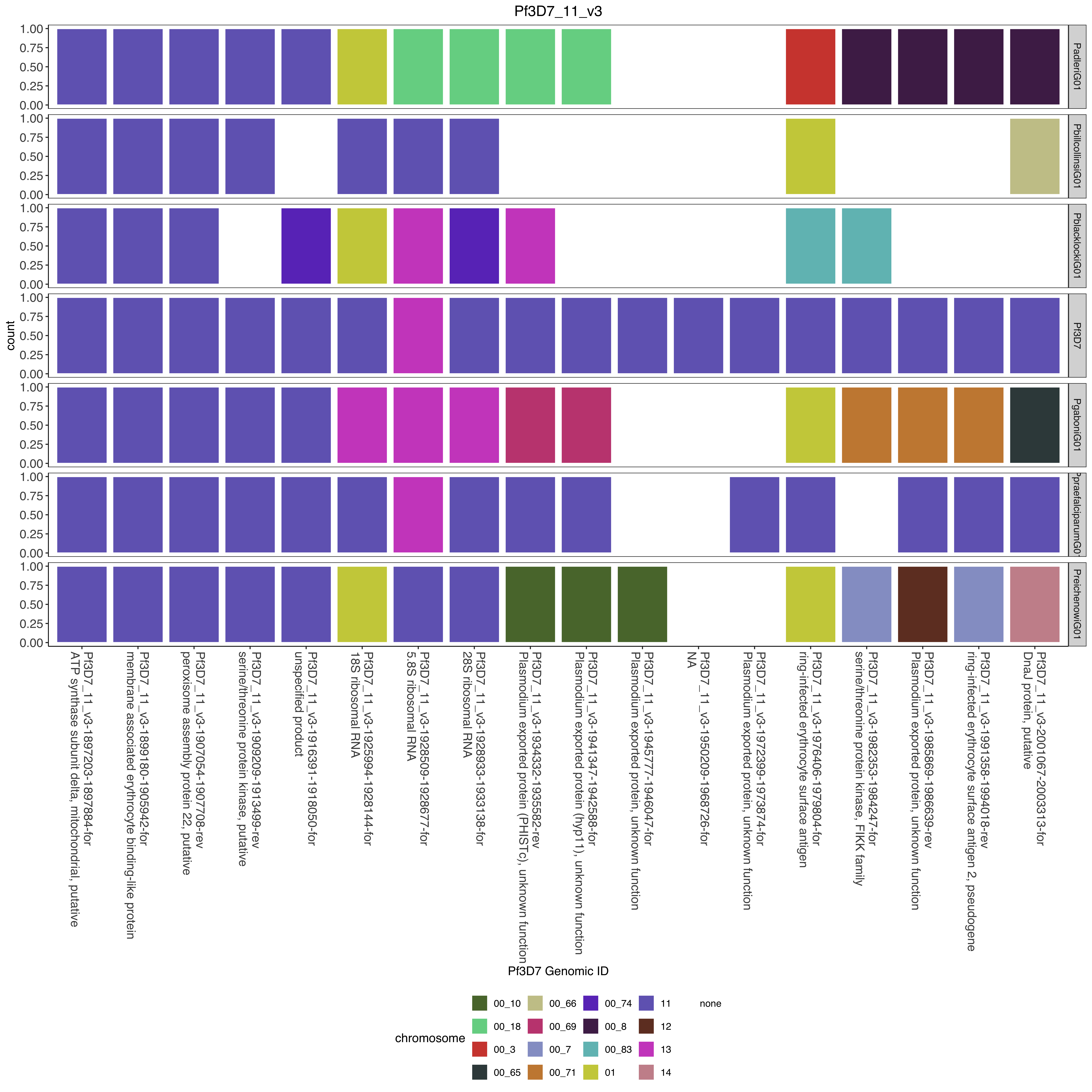
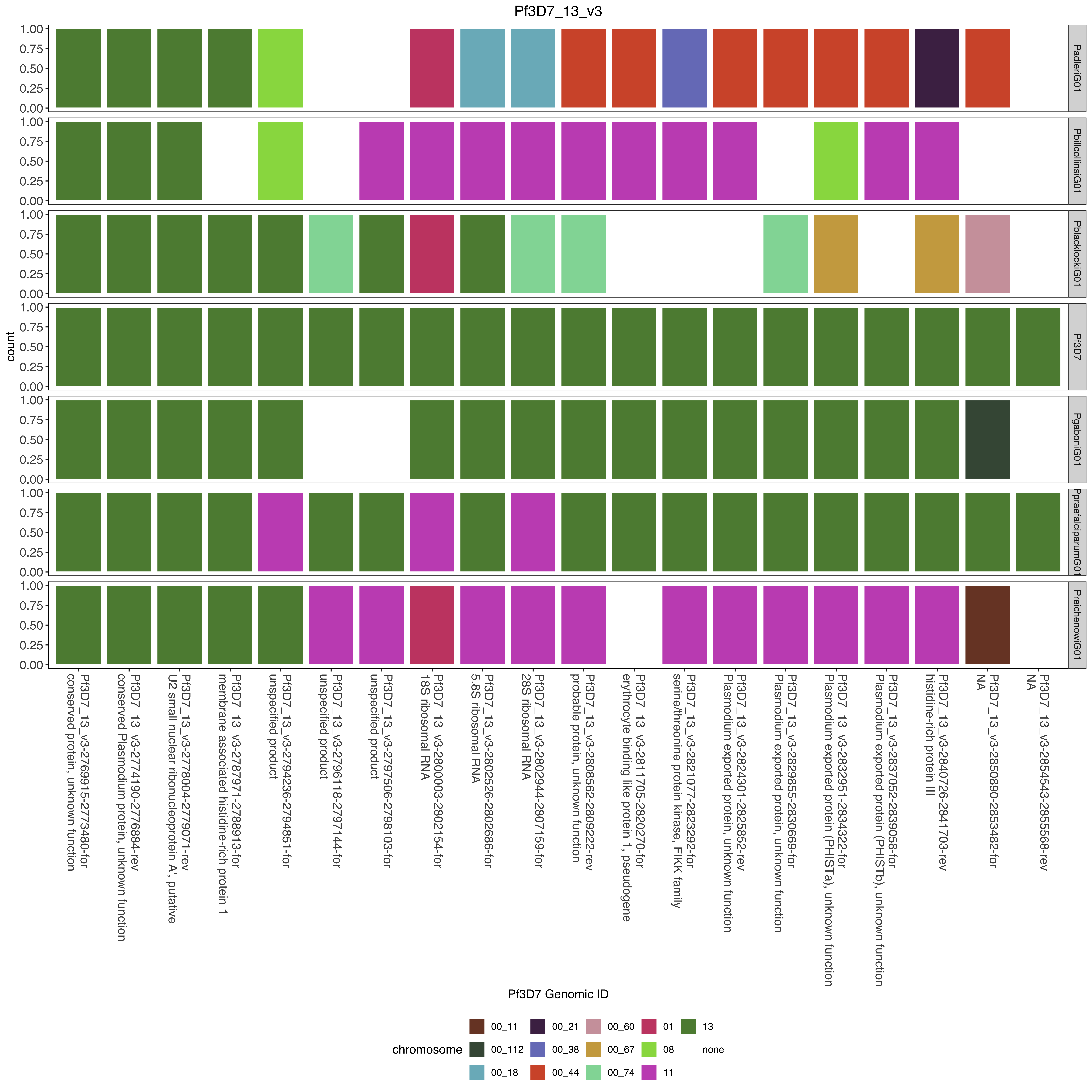
--- title: "*P Laverania* chr 05, 07, 08, 11, 13 Gene Arrangements" --- ```{r setup, echo=FALSE, message=FALSE} source("../common.R") ``` ```{bash, eval = F} elucidator gffRecordIDToGeneInfo --id PF3D7_0831800,PF3D7_1372200 --dout hrps_geneInfos --gff /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/info/gff/Pf3D7.gff --2bit /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/Pf3D7.2bit --overWriteDir elucidator extractRefSeqsFromGenomes --bed hrps_geneInfos/out_PF3D7_0831800.1.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/ --primaryGenome Pf3D7 --outputDir extractedHRPII --overWriteDirs --numThreads 7 --extendAndTrim --coverage 95 --keepBestOnly --identity 50 elucidator extractRefSeqsFromGenomes --bed hrps_geneInfos/out_PF3D7_1372200.1.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/ --primaryGenome Pf3D7 --outputDir extractedHRPIII --overWriteDirs --numThreads 7 --extendAndTrim --coverage 95 --keepBestOnly --identity 50 ``` ```{bash, eval = F} elucidator runnhmmscan --hmmModel ../MappingOutSurroundingRegions/surroundingRegionsMaterials/hmms_PfHRPs.txt --fasta /tank/data/genomes/combinedGenomes/allPLaverania/genomes/allPLaverania.fasta --overWriteDir --trimAtWhiteSpace --dout hmm_hrps_againstAllPlaverania "--defaultParameters=--nonull2 --incT 50 --incdomT 50 -T 50 --notextw --cpu 15" --hardEvalueCutOff 1e-50 ``` ```{bash, eval = F} ls /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/P*.fasta | sed 's/.fasta//g' | sed 's/.*\///g' > strains.txt elucidator gffRecordIDToGeneInfo --id PF3D7_0528800,PF3D7_0724800,PF3D7_0830300,PF3D7_1369500,PF3D7_1147700 --dout upStreamGeneInfos --gff /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/info/gff/Pf3D7.gff --2bit /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/Pf3D7.2bit --overWriteDir elucidator extractRefSeqsFromGenomes --bed upStreamGeneInfos/out_PF3D7_0528800.1.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/ --primaryGenome Pf3D7 --outputDir extractedPF3D7_0528800 --overWriteDirs --numThreads 7 --extendAndTrim elucidator extractRefSeqsFromGenomes --bed upStreamGeneInfos/out_PF3D7_0724800.1.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/ --primaryGenome Pf3D7 --outputDir extractedPF3D7_0724800 --overWriteDirs --numThreads 7 --extendAndTrim elucidator extractRefSeqsFromGenomes --bed upStreamGeneInfos/out_PF3D7_0830300.1.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/ --primaryGenome Pf3D7 --outputDir extractedPF3D7_0830300 --overWriteDirs --numThreads 7 --extendAndTrim elucidator extractRefSeqsFromGenomes --bed upStreamGeneInfos/out_PF3D7_1147700.1.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/ --primaryGenome Pf3D7 --outputDir extractedPF3D7_1147700 --overWriteDirs --numThreads 7 --extendAndTrim elucidator extractRefSeqsFromGenomes --bed upStreamGeneInfos/out_PF3D7_1369500.1.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/ --primaryGenome Pf3D7 --outputDir extractedPF3D7_1369500 --overWriteDirs --numThreads 15 --extendAndTrim mkdir chromLengths cd chromLengths for x in /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/P*.fasta; do elucidator getReadLens --fasta ${x} --trimAtWhiteSpace > $(basename ${x%%.fasta}).txt; done; cd .. mkdir endBeds for x in /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/P*.fasta; do elucidator extendToEndOfChrom --bed extractedPF3D7_0528800/Pf3D7_05_v3-1180424-1182038-rev/beds/$(basename ${x%%.fasta})_region.bed --chromLengthsTable chromLengths/$(basename ${x%%.fasta}).txt | cut -f 1-3 | elucidator bed3ToBed6 --bed STDIN --out endBeds/$(basename ${x%%.fasta})_chrom05_toEnd.bed --overWrite ; done; for x in /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/P*.fasta; do elucidator extendToEndOfChrom --bed extractedPF3D7_0724800/Pf3D7_07_v3-1047426-1049362-rev/beds/$(basename ${x%%.fasta})_region.bed --chromLengthsTable chromLengths/$(basename ${x%%.fasta}).txt | cut -f 1-3 | elucidator bed3ToBed6 --bed STDIN --out endBeds/$(basename ${x%%.fasta})_chrom07_toEnd.bed --overWrite ; done; for x in /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/P*.fasta; do elucidator extendToEndOfChrom --bed extractedPF3D7_0830300/Pf3D7_08_v3-1290239-1291406-rev/beds/$(basename ${x%%.fasta})_region.bed --chromLengthsTable chromLengths/$(basename ${x%%.fasta}).txt | cut -f 1-3 | elucidator bed3ToBed6 --bed STDIN --out endBeds/$(basename ${x%%.fasta})_chrom08_toEnd.bed --overWrite ; done; for x in /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/P*.fasta; do elucidator extendToEndOfChrom --bed extractedPF3D7_1147700/Pf3D7_11_v3-1897203-1897884-for/beds/$(basename ${x%%.fasta})_region.bed --chromLengthsTable chromLengths/$(basename ${x%%.fasta}).txt | cut -f 1-3 | elucidator bed3ToBed6 --bed STDIN --out endBeds/$(basename ${x%%.fasta})_chrom11_toEnd.bed --overWrite ; done; for x in /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/P*.fasta; do elucidator extendToEndOfChrom --bed extractedPF3D7_1369500/Pf3D7_13_v3-2769915-2773480-for/beds/$(basename ${x%%.fasta})_region.bed --chromLengthsTable chromLengths/$(basename ${x%%.fasta}).txt | cut -f 1-3 | elucidator bed3ToBed6 --bed STDIN --out endBeds/$(basename ${x%%.fasta})_chrom13_toEnd.bed --overWrite ; done; cd endBeds for x in *toEnd.bed; do elucidator gffToBedByBedLoc --extraAttributes description --feature gene,pseudogene,ncRNA_gene,protein_coding_gene --bed ${x} --gff /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/info/gff/$(echo $x | sed 's/_.*//g').gff --overWrite --out ${x%%.bed}_genes.bed; done; for x in *toEnd_genes.bed; do elucidator splitColumnContainingMeta --addHeader --delim tab --overWrite --removeEmptyColumn --column col.6 --file ${x} --out split_${x%%.bed}.tab.txt ; done; cat Pf3D7_chrom*genes.bed | elucidator splitColumnContainingMeta --file STDIN --delim tab --column col.6 --removeEmptyColumn | elucidator printCol --file STDIN --delim tab --columnName col.10 --sort --unique | egrep -v PF3D7_1148200 > allPf3D7GenesIDs.txt elucidator gffRecordIDToGeneInfo --id allPf3D7GenesIDs.txt --dout all3D7End_geneInfos --gff /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/info/gff/Pf3D7.gff --2bit /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/Pf3D7.2bit --overWriteDir elucidator getOverlappingBedRegions --bed all3D7End_geneInfos/out_allTranscripts.bed --intersectWithBed ../../windowAnalysis/windows/finalHRPII_HRPIII_windows_withTuned.bed --overWrite | egrep -v Pf3D7_11_v3_1972399_1973874_PF3D7_1149100.2 | egrep -v Pf3D7_11_v3_1899180_1905695_PF3D7_1147800.2 | egrep -v Pf3D7_11_v3_1950209_1968726_PF3D7_1149000.1 > all3D7End_geneInfos/selected_out_allTranscripts.bed egrep -i ribosomal Pf3D7_chrom11_toEnd_genes.bed >> all3D7End_geneInfos/selected_out_allTranscripts.bed egrep -i ribosomal Pf3D7_chrom13_toEnd_genes.bed >> all3D7End_geneInfos/selected_out_allTranscripts.bed elucidator extractRefSeqsFromGenomes --bed all3D7End_geneInfos/selected_out_allTranscripts.bed --genomeDir /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/ --primaryGenome Pf3D7 --outputDir extractedPF3D7_endGenes --overWriteDirs --numThreads 20 --extendAndTrim --keepBestOnly cd extractedPF3D7_endGenes for x in Pf3D7_*/beds/*bestRegion.bed; do elucidator addColumn --file ${x} --newColumnName Pf3D7_genomicID --element $(echo ${x} | sed 's/\/.*//g') --overWrite --out ${x%%.bed}_withGenomcId.bed --delim tab; done; for x in Pf3D7_*/beds/*bestRegion_withGenomcId.bed; do elucidator addColumn --file ${x} --newColumnName strain --element $(echo ${x} | sed 's/.*\///g' | sed 's/_.*//g') --overWrite --out ${x%%.bed}_withStrain.bed --delim tab; done; /bin/ls Pf3D7_*/beds/*bestRegion_withGenomcId_withStrain.bed > allBestRegionsFiles.txt elucidator rBind --files allBestRegionsFiles.txt --delim tab --overWrite --out allBestRegions.bed ``` # Finding possible fragments in extra contigs ```{r} = readr:: read_tsv ("strains.txt" , col_names = c ("strains" )) = readr:: read_tsv ("endBeds/all3D7End_geneInfos/selected_out_allTranscripts.bed" , col_names = F) %>% mutate (genomicID = paste0 (X1, "-" , X2, "-" , X3, "-" , ifelse (X6 == "-" , "rev" , "for" ))) %>% mutate (` Pf3D7_genomicID ` = genomicID) %>% mutate (` Pf3D7_GeneID ` = gsub (".*ID=" , "" , gsub (";.*" , "" , X7))) %>% mutate (` Pf3D7_GeneDescription ` = gsub (".*description=" , "" , gsub (";feature.*" , "" , X7))) %>% mutate (` Pf3D7_GeneID ` = ifelse (grepl ("ribosomal RNA" , X7), gsub (";.*" , "" , gsub (".*;ID=" , "" , X7)), Pf3D7_GeneID))%>% mutate (` Pf3D7_GeneDescription ` = ifelse (grepl ("ribosomal RNA" , X7), gsub (";.*" , "" , gsub (".*;description=" , "" , X7)), Pf3D7_GeneDescription))= readr:: read_tsv ("endBeds/extractedPF3D7_endGenes/allBestRegions.bed" , col_names = F) %>% rename (Pf3D7_genomicID = X7, strain = X8)``` # Detecting the prescence of genes using blast ```{r} = allStrainLocs %>% complete (Pf3D7_genomicID, strain) %>% left_join (Pf3d7_endRegions %>% select (starts_with ("Pf3D7" ))) %>% mutate (Pf3D7_chrom = gsub ("-.*" , "" , Pf3D7_genomicID)) %>% replace_na (list (X1 = "none" ))pdf ("3D7_geneLocations_in_lstrains_byStrain.pdf" , width = 15 , height = 20 , useDingbats = F)for (lstrain in strains$ strains) {= allStrainLocsMod %>% filter (strain == lstrain)= c (scheme$ hex (length (unique (allStrainLocsMod_strain$ X1))))names (chromHitColors) = unique (allStrainLocsMod_strain$ X1)"none" ] = "white" print (ggplot (allStrainLocsMod_strain) + geom_bar (aes (x = paste0 (Pf3D7_genomicID, " \n " , Pf3D7_GeneDescription),fill = X1+ facet_wrap (~ Pf3D7_chrom, strip.position = "right" ,ncol = 1 , scales = "free" ) + scale_fill_manual (values = chromHitColors) + + labs (title = lstrain, x = "Pf3D7 Genomic ID" ))dev.off ()``` ## Displayed by strain ```{r} #| fig-column: screen-inset-shaded #| fig-height: 20 for (lstrain in strains$ strains) {= allStrainLocsMod %>% filter (strain == lstrain)= c (scheme$ hex (length (unique (allStrainLocsMod_strain$ X1))))names (chromHitColors) = unique (allStrainLocsMod_strain$ X1)"none" ] = "white" print (ggplot (allStrainLocsMod_strain) + geom_bar (aes (x = paste0 (Pf3D7_genomicID, " \n " , Pf3D7_GeneDescription),fill = X1+ facet_wrap (~ Pf3D7_chrom,strip.position = "right" , ncol = 1 , scales = "free" ) + scale_fill_manual (values = chromHitColors) + + labs (title = lstrain, x = "Pf3D7 Genomic ID" ))``` ```{r} pdf ("3D7_geneLocations_in_lstrains_byChrom.pdf" , width = 15 , height = 20 , useDingbats = F)for (chrom in unique (allStrainLocsMod$ Pf3D7_chrom)) {= allStrainLocsMod %>% filter (Pf3D7_chrom == chrom) %>% mutate (strainChromNumber = sub (".*?_" , "" , X1)) %>% mutate (strainChromNumber = sub ("_v3" , "" , strainChromNumber))= c (scheme$ hex (length (unique (allStrainLocsMod_chrom$ strainChromNumber))))names (chromHitColors) = unique (allStrainLocsMod_chrom$ strainChromNumber)"none" ] = "white" print (ggplot (allStrainLocsMod_chrom) + geom_bar (aes (x = paste0 (Pf3D7_genomicID, " \n " , Pf3D7_GeneDescription),fill = strainChromNumbercolor = "white" ) + facet_wrap (~ strain, strip.position = "right" , ncol = 1 ) + scale_fill_manual (values = chromHitColors) + + labs (title = chrom, x = "Pf3D7 Genomic ID" , fill = "chromosome" ))dev.off ()``` ## Displayed by chromsome ```{r} #| fig-column: screen-inset-shaded #| fig-width: 15 #| fig-height: 15 for (chrom in unique (allStrainLocsMod$ Pf3D7_chrom)) {= allStrainLocsMod %>% filter (Pf3D7_chrom == chrom) %>% mutate (strainChromNumber = sub (".*?_" , "" , X1)) %>% mutate (strainChromNumber = sub ("_v3" , "" , strainChromNumber))= c (scheme$ hex (length (unique (allStrainLocsMod_chrom$ strainChromNumber))))names (chromHitColors) = unique (allStrainLocsMod_chrom$ strainChromNumber)"none" ] = "white" print (ggplot (allStrainLocsMod_chrom) + geom_bar (aes (x = paste0 (Pf3D7_genomicID, " \n " , Pf3D7_GeneDescription),fill = strainChromNumbercolor = "white" ) + facet_wrap (~ strain, strip.position = "right" , ncol = 1 ) + scale_fill_manual (values = chromHitColors) + + labs (title = chrom, x = "Pf3D7 Genomic ID" , fill = "chromosome" ))``` ## finding possible fragments of 8 ```{bash, eval = F} elucidator fastaToBed --trimAtWhiteSpace --fasta /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/PreichenowiG01.fasta | egrep PRG01_00_67 | elucidator bed3ToBed6 --bed STDIN --out PRG01_00_67.bed --overWrite elucidator gffToBedByBedLoc --extraAttributes description --feature gene,pseudogene,ncRNA_gene,protein_coding_gene --bed PRG01_00_67.bed --gff /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/info/gff/PreichenowiG01.gff --overWrite --out PRG01_00_67_genes.bed elucidator fastaToBed --trimAtWhiteSpace --fasta /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/PadleriG01.fasta | egrep PADLG01_00_21 | elucidator bed3ToBed6 --bed STDIN --out PADLG01_00_21.bed --overWrite elucidator gffToBedByBedLoc --extraAttributes description --feature gene,pseudogene,ncRNA_gene,protein_coding_gene --bed PADLG01_00_21.bed --gff /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/info/gff/PadleriG01.gff --overWrite --out PADLG01_00_21_genes.bed echo -e "PGABG01_08\t0\t10000\tPRG01_08-0-10000\t10000\t+" > PGABG01_08_front.bed elucidator gffToBedByBedLoc --extraAttributes description --feature gene,pseudogene,ncRNA_gene,protein_coding_gene --bed PGABG01_08_front.bed --gff /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/info/gff/PgaboniG01.gff --overWrite --out PGABG01_08_front_genes.bed for x in *_genes.bed; do elucidator splitColumnContainingMeta --addHeader --delim tab --overWrite --removeEmptyColumn --column col.6 --file ${x} --out split_${x%%.bed}.tab.txt ; done; ``` ## finding possible fragments of 11 ```{bash, eval = F} PGABG01_00_71,PGABG01_00_69 elucidator fastaToBed --trimAtWhiteSpace --fasta /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/PgaboniG01.fasta | egrep PGABG01_00_71 | elucidator bed3ToBed6 --bed STDIN --out PGABG01_00_71.bed --overWrite elucidator gffToBedByBedLoc --extraAttributes description --feature gene,pseudogene,ncRNA_gene,protein_coding_gene --bed PGABG01_00_71.bed --gff /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/info/gff/PgaboniG01.gff --overWrite --out PGABG01_00_71_genes.bed elucidator fastaToBed --trimAtWhiteSpace --fasta /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/PgaboniG01.fasta | egrep PGABG01_00_69 | elucidator bed3ToBed6 --bed STDIN --out PGABG01_00_69.bed --overWrite elucidator gffToBedByBedLoc --extraAttributes description --feature gene,pseudogene,ncRNA_gene,protein_coding_gene --bed PGABG01_00_69.bed --gff /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/info/gff/PgaboniG01.gff --overWrite --out PGABG01_00_69_genes.bed ``` ## finding possible fragments of 13 ```{bash, eval = F} #PADLG01_00_60 just EBL-1 fragment #PADLG01_00_44 elucidator fastaToBed --trimAtWhiteSpace --fasta /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/genomes/PadleriG01.fasta | egrep PADLG01_00_44 | elucidator bed3ToBed6 --bed STDIN --out PADLG01_00_44.bed --overWrite elucidator gffToBedByBedLoc --extraAttributes description --feature gene,pseudogene,ncRNA_gene,protein_coding_gene --bed PADLG01_00_44.bed --gff /tank/data/genomes/plasmodium/genomes/pLaveraniaPlus3D7/info/gff/PadleriG01.gff --overWrite --out PADLG01_00_44_genes.bed ``` # Reading In Genes ```{r} = readr:: read_tsv ("strains.txt" , col_names = c ("strains" )) = tibble ()for (strain in strains$ strains){= readr:: read_tsv (paste0 ("chromLengths/" , strain, ".txt" ), col_names = c ("chrom" , "length" )) %>% mutate (strain = strain)= bind_rows (chromLengths, strainLens)``` ## 05 ```{r} #| code-fold: true = tibble ()for (strain in strains$ strains){= readr:: read_tsv (paste0 ("endBeds/split_" , strain, "_chrom05_toEnd_genes.tab.txt" )) %>% mutate (strain = strain ) %>% mutate (chromGlobal = gsub (paste0 (strain, "_" ), "" , col.0 )) %>% mutate (chromGlobal = gsub ("_v3" , "" , chromGlobal)) %>% rename (chrom = col.0 , start = col.1 , end = col.2 )= bind_rows (all05, strain05)= all05 %>% mutate (description = gsub ("unknownfunction" , "unknown function" , description)) %>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse ("membrane associated erythrocyte binding-like protein" == description, "merozoite adhesive erythrocytic binding protein" , description)) %>% mutate (description = ifelse ("membrane associated erythrocyte binding-likeprotein" == description, "merozoite adhesive erythrocytic binding protein" , description)) %>% mutate (description = ifelse ("membrane associated histidine-rich protein 1" == description, "membrane associated histidine-rich protein" , description)) %>% mutate (description = gsub (",putative" , ", putative" , description)) %>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description))%>% mutate (description = gsub ("unknownfunction" , "unknown function" , description))%>% mutate (description = gsub ("conserved protein, unknown function" , "conserved Plasmodium protein, unknown function" , description)) %>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse ("membrane associated histidine-rich protein 1" == description, "membrane associated histidine-rich protein" , description))%>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description))%>% mutate (description = gsub ("unknownfunction" , "unknown function" , description))%>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse (grepl ("sporozoite and liver stage tryptophan-rich protein, putative" , description), "tryptophan/threonine-rich antigen" , description))%>% mutate (description = ifelse (grepl ("CRA domain-containing protein, putative" , description), "conserved Plasmodium protein, unknown function" , description))%>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description)) %>% mutate (description = gsub ("surfaceantigen" , "surface antigen" , description)) %>% mutate (description = gsub ("Tetratricopeptide repeat, putative" , "tetratricopeptide repeat protein, putative" , description)) %>% mutate (description = gsub ("transmembraneprotein" , "transmembrane protein" , description)) %>% mutate (description = ifelse (grepl ("PfEMP1" , description) & grepl ("pseudogene" , description), "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description)) %>% mutate (description = gsub ("PIR protein" , "stevor" , description)) %>% mutate (description = gsub ("erythrocyte membrane protein 1-like" , "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description)) %>% mutate (description = gsub ("acidic terminal segments, variant surface antigen of PfEMP1, putative" , "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description))%>% mutate (description = ifelse (grepl ("CoA binding protein" , description, ignore.case = T), "acyl-CoA binding protein" , description)) %>% mutate (description = ifelse (grepl ("transfer RNA" , description) | grepl ("tRNA" , description), "tRNA" , description))%>% mutate (description = ifelse (grepl ("cytoadherence" , description), "CLAG" , description))%>% mutate (description = ifelse (grepl ("surface-associated interspersed protein" , description), "SURFIN" , description))%>% mutate (description = ifelse (grepl ("SURFIN" , description), "SURFIN" , description))%>% mutate (description = ifelse (grepl ("stevor-like" , description), "stevor, pseudogene" , description)) %>% mutate (description = ifelse (grepl ("exported protein family" , description), "exported protein family" , description)) %>% mutate (description = ifelse (grepl ("ribosomal RNA" , description), "rRNA" , description)) %>% mutate (description = ifelse (grepl ("serine/threonine protein kinase" , description), "serine/threonine protein kinase, FIKK family" , description)) %>% mutate (description = ifelse (grepl ("hypothetical protein" , description), "hypothetical protein, conserved" , description)) %>% mutate (description = ifelse (grepl ("conserved Plasmodium protein, unknown function" , description), "hypothetical protein, conserved" , description)) %>% mutate (description = ifelse (grepl ("Rifin/stevor family, putative" , description), "stevor" , description)) %>% mutate (description = ifelse (grepl ("stevor" , description), "stevor" , description)) %>% mutate (description = ifelse (grepl ("rifin" , description), "rifin" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description), "erythrocyte membrane protein 1 (PfEMP1)" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte membrane protein 1" , description), "erythrocyte membrane protein 1 (PfEMP1)" , description)) %>% mutate (description = ifelse (grepl ("probably protein" , description), "unspecified product" , description)) %>% mutate (description = ifelse (grepl ("RESA" , description), "RESA" , description)) %>% mutate (description = ifelse (grepl ("ring-infected erythrocyte surface antigen" , description), "ring-infected erythrocyte surface antigen" , description)) %>% mutate (description = ifelse (grepl ("Duffy binding domain/Erythrocyte binding antigen175, putative" , description), "erythrocyte binding like protein 1" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte binding like protein 1" , description), "erythrocyte binding like protein 1" , description)) %>% mutate (description = ifelse (grepl ("unspecified product" , description), "hypothetical protein, conserved" , description))%>% mutate (description = ifelse (grepl ("probable protein, unknown function" , description), "hypothetical protein, conserved" , description))%>% mutate (description = ifelse (grepl ("RESA" , description), "ring-infected erythrocyte surface antigen" , description)) %>% mutate (description = gsub (", putative" , "" , description))= all05 %>% group_by (strain, chrom) %>% summarise (minStart = min (start))= all05 %>% left_join (all05_strainStarts) %>% mutate (globalStart = start - minStart, globalEnd = end - minStart)= chromLengths %>% filter (chrom %in% all05_strainStarts$ chrom) %>% left_join (all05_strainStarts)%>% mutate (globalStart = 0 , globalEnd = length - minStart) %>% mutate (chrom = factor (chrom, levels = chrom))= all05 %>% mutate (chrom = factor (chrom, levels = chromLengths_05$ chrom))``` ## 07 ```{r} #| code-fold: true = tibble ()for (strain in strains$ strains){= readr:: read_tsv (paste0 ("endBeds/split_" , strain, "_chrom07_toEnd_genes.tab.txt" )) %>% mutate (strain = strain ) %>% mutate (chromGlobal = gsub (paste0 (strain, "_" ), "" , col.0 )) %>% mutate (chromGlobal = gsub ("_v3" , "" , chromGlobal)) %>% rename (chrom = col.0 , start = col.1 , end = col.2 )= bind_rows (all07, strain07)= all07 %>% mutate (description = gsub ("unknownfunction" , "unknown function" , description)) %>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse ("membrane associated erythrocyte binding-like protein" == description, "merozoite adhesive erythrocytic binding protein" , description)) %>% mutate (description = ifelse ("membrane associated erythrocyte binding-likeprotein" == description, "merozoite adhesive erythrocytic binding protein" , description)) %>% mutate (description = ifelse ("membrane associated histidine-rich protein 1" == description, "membrane associated histidine-rich protein" , description)) %>% mutate (description = gsub (",putative" , ", putative" , description)) %>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description))%>% mutate (description = gsub ("unknownfunction" , "unknown function" , description))%>% mutate (description = gsub ("conserved protein, unknown function" , "conserved Plasmodium protein, unknown function" , description)) %>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse ("membrane associated histidine-rich protein 1" == description, "membrane associated histidine-rich protein" , description))%>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description))%>% mutate (description = gsub ("unknownfunction" , "unknown function" , description))%>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse (grepl ("sporozoite and liver stage tryptophan-rich protein, putative" , description), "tryptophan/threonine-rich antigen" , description))%>% mutate (description = ifelse (grepl ("CRA domain-containing protein, putative" , description), "conserved Plasmodium protein, unknown function" , description))%>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description)) %>% mutate (description = gsub ("surfaceantigen" , "surface antigen" , description)) %>% mutate (description = gsub ("Tetratricopeptide repeat, putative" , "tetratricopeptide repeat protein, putative" , description)) %>% mutate (description = gsub ("transmembraneprotein" , "transmembrane protein" , description)) %>% mutate (description = ifelse (grepl ("PfEMP1" , description) & grepl ("pseudogene" , description), "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description)) %>% mutate (description = gsub ("PIR protein" , "stevor" , description)) %>% mutate (description = gsub ("erythrocyte membrane protein 1-like" , "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description)) %>% mutate (description = gsub ("acidic terminal segments, variant surface antigen of PfEMP1, putative" , "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description))%>% mutate (description = ifelse (grepl ("CoA binding protein" , description, ignore.case = T), "acyl-CoA binding protein" , description)) %>% mutate (description = ifelse (grepl ("transfer RNA" , description) | grepl ("tRNA" , description), "tRNA" , description))%>% mutate (description = ifelse (grepl ("cytoadherence" , description), "CLAG" , description))%>% mutate (description = ifelse (grepl ("surface-associated interspersed protein" , description), "SURFIN" , description))%>% mutate (description = ifelse (grepl ("SURFIN" , description), "SURFIN" , description))%>% mutate (description = ifelse (grepl ("stevor-like" , description), "stevor, pseudogene" , description)) %>% mutate (description = ifelse (grepl ("exported protein family" , description), "exported protein family" , description)) %>% mutate (description = ifelse (grepl ("ribosomal RNA" , description), "rRNA" , description)) %>% mutate (description = ifelse (grepl ("serine/threonine protein kinase" , description), "serine/threonine protein kinase, FIKK family" , description)) %>% mutate (description = ifelse (grepl ("hypothetical protein" , description), "hypothetical protein, conserved" , description)) %>% mutate (description = ifelse (grepl ("conserved Plasmodium protein, unknown function" , description), "hypothetical protein, conserved" , description)) %>% mutate (description = ifelse (grepl ("Rifin/stevor family, putative" , description), "stevor" , description)) %>% mutate (description = ifelse (grepl ("stevor" , description), "stevor" , description)) %>% mutate (description = ifelse (grepl ("rifin" , description), "rifin" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description), "erythrocyte membrane protein 1 (PfEMP1)" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte membrane protein 1" , description), "erythrocyte membrane protein 1 (PfEMP1)" , description)) %>% mutate (description = ifelse (grepl ("probably protein" , description), "unspecified product" , description)) %>% mutate (description = ifelse (grepl ("RESA" , description), "RESA" , description)) %>% mutate (description = ifelse (grepl ("ring-infected erythrocyte surface antigen" , description), "ring-infected erythrocyte surface antigen" , description)) %>% mutate (description = ifelse (grepl ("Duffy binding domain/Erythrocyte binding antigen175, putative" , description), "erythrocyte binding like protein 1" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte binding like protein 1" , description), "erythrocyte binding like protein 1" , description)) %>% mutate (description = ifelse (grepl ("unspecified product" , description), "hypothetical protein, conserved" , description))%>% mutate (description = ifelse (grepl ("probable protein, unknown function" , description), "hypothetical protein, conserved" , description)) %>% mutate (description = ifelse (grepl ("RESA" , description), "ring-infected erythrocyte surface antigen" , description)) %>% mutate (description = gsub (", putative" , "" , description)) %>% mutate (description = ifelse (grepl ("transcription factor with AP2 domain(s)" , description, fixed = T), "AP2 domain transcription factor AP2-L" , description)) %>% mutate (description = ifelse (grepl ("zinc finger, C3HC4 type" , description, fixed = T), "RING zinc finger protein" , description)) %>% mutate (description = ifelse (grepl ("eukaryotic translation initiation factor 2 subunit alpha" , description, fixed = T), "eukaryotic translation initiation factor 2 alpha subunit" , description)) = all07 %>% group_by (strain, chrom) %>% summarise (minStart = min (start))= all07 %>% left_join (all07_strainStarts) %>% mutate (globalStart = start - minStart, globalEnd = end - minStart)= chromLengths %>% filter (chrom %in% all07_strainStarts$ chrom) %>% left_join (all07_strainStarts)%>% mutate (globalStart = 0 , globalEnd = length - minStart) %>% mutate (chrom = factor (chrom, levels = chrom))= all07 %>% mutate (chrom = factor (chrom, levels = chromLengths_07$ chrom))``` ## 08 ```{r} #| code-fold: true = tibble ()for (strain in strains$ strains){= readr:: read_tsv (paste0 ("endBeds/split_" , strain, "_chrom08_toEnd_genes.tab.txt" )) %>% mutate (strain = strain ) %>% mutate (chromGlobal = gsub (paste0 (strain, "_" ), "" , col.0 )) %>% mutate (chromGlobal = gsub ("_v3" , "" , chromGlobal)) %>% rename (chrom = col.0 , start = col.1 , end = col.2 )= bind_rows (all08, strain08)= bind_rows (:: read_tsv ("possibleFragments/split_PRG01_00_67_genes.tab.txt" ), :: read_tsv ("possibleFragments/split_PADLG01_00_21_genes.tab.txt" ), :: read_tsv ("possibleFragments/split_PGABG01_08_front_genes.tab.txt" )%>% mutate (chromGlobal = gsub (paste0 (strain, "_" ), "" , col.0 )) %>% mutate (chromGlobal = gsub ("_v3" , "" , chromGlobal)) %>% rename (chrom = col.0 , start = col.1 , end = col.2 )= all08 %>% mutate (description = gsub ("unknownfunction" , "unknown function" , description)) %>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse ("membrane associated erythrocyte binding-like protein" == description, "merozoite adhesive erythrocytic binding protein" , description)) %>% mutate (description = ifelse ("membrane associated erythrocyte binding-likeprotein" == description, "merozoite adhesive erythrocytic binding protein" , description)) %>% mutate (description = ifelse ("membrane associated histidine-rich protein 1" == description, "membrane associated histidine-rich protein" , description)) %>% mutate (description = gsub (",putative" , ", putative" , description)) %>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description))%>% mutate (description = gsub ("unknownfunction" , "unknown function" , description))%>% mutate (description = gsub ("conserved protein, unknown function" , "conserved Plasmodium protein, unknown function" , description)) %>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse ("membrane associated histidine-rich protein 1" == description, "membrane associated histidine-rich protein" , description))%>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description))%>% mutate (description = gsub ("unknownfunction" , "unknown function" , description))%>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse (grepl ("sporozoite and liver stage tryptophan-rich protein, putative" , description), "tryptophan/threonine-rich antigen" , description))%>% mutate (description = ifelse (grepl ("CRA domain-containing protein, putative" , description), "conserved Plasmodium protein, unknown function" , description))%>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description)) %>% mutate (description = gsub ("surfaceantigen" , "surface antigen" , description)) %>% mutate (description = gsub ("Tetratricopeptide repeat, putative" , "tetratricopeptide repeat protein, putative" , description)) %>% mutate (description = gsub ("transmembraneprotein" , "transmembrane protein" , description)) %>% mutate (description = ifelse (grepl ("PfEMP1" , description) & grepl ("pseudogene" , description), "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description)) %>% mutate (description = gsub ("PIR protein" , "stevor" , description)) %>% mutate (description = gsub ("erythrocyte membrane protein 1-like" , "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description)) %>% mutate (description = gsub ("acidic terminal segments, variant surface antigen of PfEMP1, putative" , "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description))%>% mutate (description = ifelse (grepl ("CoA binding protein" , description, ignore.case = T), "acyl-CoA binding protein" , description)) %>% mutate (description = ifelse (grepl ("transfer RNA" , description) | grepl ("tRNA" , description), "tRNA" , description))%>% mutate (description = ifelse (grepl ("cytoadherence" , description), "CLAG" , description))%>% mutate (description = ifelse (grepl ("surface-associated interspersed protein" , description), "SURFIN" , description))%>% mutate (description = ifelse (grepl ("SURFIN" , description), "SURFIN" , description))%>% mutate (description = ifelse (grepl ("stevor-like" , description), "stevor, pseudogene" , description)) %>% mutate (description = ifelse (grepl ("exported protein family" , description), "exported protein family" , description)) %>% mutate (description = ifelse (grepl ("ribosomal RNA" , description), "rRNA" , description)) %>% mutate (description = ifelse (grepl ("serine/threonine protein kinase" , description), "serine/threonine protein kinase, FIKK family" , description)) %>% mutate (description = ifelse (grepl ("hypothetical protein" , description), "hypothetical protein, conserved" , description)) %>% mutate (description = ifelse (grepl ("conserved Plasmodium protein, unknown function" , description), "hypothetical protein, conserved" , description)) %>% mutate (description = ifelse (grepl ("Rifin/stevor family, putative" , description), "stevor" , description)) %>% mutate (description = ifelse (grepl ("stevor" , description), "stevor" , description)) %>% mutate (description = ifelse (grepl ("rifin" , description), "rifin" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description), "erythrocyte membrane protein 1 (PfEMP1)" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte membrane protein 1" , description), "erythrocyte membrane protein 1 (PfEMP1)" , description)) %>% mutate (description = ifelse (grepl ("probably protein" , description), "unspecified product" , description)) %>% mutate (description = ifelse (grepl ("RESA" , description), "RESA" , description)) %>% mutate (description = ifelse (grepl ("ring-infected erythrocyte surface antigen" , description), "ring-infected erythrocyte surface antigen" , description)) %>% mutate (description = ifelse (grepl ("Duffy binding domain/Erythrocyte binding antigen175, putative" , description), "erythrocyte binding like protein 1" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte binding like protein 1" , description), "erythrocyte binding like protein 1" , description)) %>% mutate (description = ifelse (grepl ("unspecified product" , description), "hypothetical protein, conserved" , description))%>% mutate (description = ifelse (grepl ("probable protein, unknown function" , description), "hypothetical protein, conserved" , description))%>% mutate (description = ifelse (grepl ("RESA" , description), "ring-infected erythrocyte surface antigen" , description)) %>% mutate (description = gsub (", putative" , "" , description))= all08 %>% group_by (strain, chrom) %>% summarise (minStart = min (start))= all08 %>% left_join (all08_strainStarts) %>% mutate (globalStart = start - minStart, globalEnd = end - minStart)= chromLengths %>% filter (chrom %in% all08_strainStarts$ chrom) %>% left_join (all08_strainStarts)%>% mutate (globalStart = 0 , globalEnd = length - minStart) %>% mutate (chrom = factor (chrom, levels = chrom))= all08 %>% mutate (chrom = factor (chrom, levels = chromLengths_08$ chrom))``` ## 13 ```{r} #| code-fold: true = tibble ()for (strain in strains$ strains){= readr:: read_tsv (paste0 ("endBeds/split_" , strain, "_chrom13_toEnd_genes.tab.txt" )) %>% mutate (strain = strain ) %>% mutate (chromGlobal = gsub (paste0 (strain, "_" ), "" , col.0 )) %>% mutate (chromGlobal = gsub ("_v3" , "" , chromGlobal)) %>% rename (chrom = col.0 , start = col.1 , end = col.2 )= bind_rows (all13, strain13)= all13 %>% filter (col.3 %!in% c ("Pf3D7_13_v3_2794236_2794851_PF3D7_1370500" , "Pf3D7_13_v3_2796118_2797144_PF3D7_1370800" , "Pf3D7_13_v3_2797506_2798103_PF3D7_1370900" )) %>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse ("membrane associated erythrocyte binding-like protein" == description, "merozoite adhesive erythrocytic binding protein" , description)) %>% mutate (description = ifelse ("membrane associated erythrocyte binding-likeprotein" == description, "merozoite adhesive erythrocytic binding protein" , description)) %>% mutate (description = ifelse ("membrane associated histidine-rich protein 1" == description, "membrane associated histidine-rich protein" , description)) %>% mutate (description = gsub (",putative" , ", putative" , description)) %>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description))%>% mutate (description = gsub ("unknownfunction" , "unknown function" , description))%>% mutate (description = gsub ("conserved protein, unknown function" , "conserved Plasmodium protein, unknown function" , description)) %>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse ("membrane associated histidine-rich protein 1" == description, "membrane associated histidine-rich protein" , description))%>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description))%>% mutate (description = gsub ("unknownfunction" , "unknown function" , description))%>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse (grepl ("sporozoite and liver stage tryptophan-rich protein, putative" , description), "tryptophan/threonine-rich antigen" , description))%>% mutate (description = ifelse (grepl ("CRA domain-containing protein, putative" , description), "conserved Plasmodium protein, unknown function" , description))%>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description)) %>% mutate (description = gsub ("surfaceantigen" , "surface antigen" , description)) %>% mutate (description = gsub ("Tetratricopeptide repeat, putative" , "tetratricopeptide repeat protein, putative" , description)) %>% mutate (description = gsub ("transmembraneprotein" , "transmembrane protein" , description)) %>% mutate (description = ifelse (grepl ("PfEMP1" , description) & grepl ("pseudogene" , description), "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description)) %>% mutate (description = gsub ("PIR protein" , "stevor" , description)) %>% mutate (description = gsub ("erythrocyte membrane protein 1-like" , "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description)) %>% mutate (description = gsub ("acidic terminal segments, variant surface antigen of PfEMP1, putative" , "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description))%>% mutate (description = ifelse (grepl ("CoA binding protein" , description, ignore.case = T), "acyl-CoA binding protein" , description)) %>% mutate (description = ifelse (grepl ("transfer RNA" , description) | grepl ("tRNA" , description), "tRNA" , description))%>% mutate (description = ifelse (grepl ("cytoadherence" , description), "CLAG" , description))%>% mutate (description = ifelse (grepl ("surface-associated interspersed protein" , description), "SURFIN" , description))%>% mutate (description = ifelse (grepl ("SURFIN" , description), "SURFIN" , description))%>% mutate (description = ifelse (grepl ("stevor-like" , description), "stevor, pseudogene" , description)) %>% mutate (description = ifelse (grepl ("exported protein family" , description), "exported protein family" , description)) %>% mutate (description = ifelse (grepl ("ribosomal RNA" , description), "rRNA" , description)) %>% mutate (description = ifelse (grepl ("serine/threonine protein kinase" , description), "serine/threonine protein kinase, FIKK family" , description)) %>% mutate (description = ifelse (grepl ("hypothetical protein" , description), "hypothetical protein, conserved" , description)) %>% mutate (description = ifelse (grepl ("conserved Plasmodium protein, unknown function" , description), "hypothetical protein, conserved" , description)) %>% mutate (description = ifelse (grepl ("Rifin/stevor family, putative" , description), "stevor" , description)) %>% mutate (description = ifelse (grepl ("stevor" , description), "stevor" , description)) %>% mutate (description = ifelse (grepl ("rifin" , description), "rifin" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description), "erythrocyte membrane protein 1 (PfEMP1)" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte membrane protein 1" , description), "erythrocyte membrane protein 1 (PfEMP1)" , description)) %>% mutate (description = ifelse (grepl ("probably protein" , description), "unspecified product" , description)) %>% mutate (description = ifelse (grepl ("RESA" , description), "RESA" , description)) %>% mutate (description = ifelse (grepl ("ring-infected erythrocyte surface antigen" , description), "ring-infected erythrocyte surface antigen" , description))%>% mutate (description = ifelse (grepl ("Duffy binding domain/Erythrocyte binding antigen175, putative" , description), "erythrocyte binding like protein 1" , description))%>% mutate (description = ifelse (grepl ("erythrocyte binding like protein 1" , description), "erythrocyte binding like protein 1" , description)) %>% mutate (description = ifelse (grepl ("unspecified product" , description), "hypothetical protein, conserved" , description))%>% mutate (description = ifelse (grepl ("probable protein, unknown function" , description), "hypothetical protein, conserved" , description))%>% mutate (description = ifelse (grepl ("RESA" , description), "ring-infected erythrocyte surface antigen" , description)) %>% mutate (description = gsub (", putative" , "" , description))= all13 %>% group_by (strain, chrom) %>% summarise (minStart = min (start))= all13 %>% left_join (all13_strainStarts) %>% mutate (globalStart = start - minStart, globalEnd = end - minStart)= chromLengths %>% filter (chrom %in% all13_strainStarts$ chrom) %>% left_join (all13_strainStarts)%>% mutate (globalStart = 0 , globalEnd = length - minStart) %>% mutate (chrom = factor (chrom, levels = chrom))= all13 %>% mutate (chrom = factor (chrom, levels = chromLengths_13$ chrom))``` ## 11 ```{r} #| code-fold: true = tibble ()for (strain in strains$ strains){= readr:: read_tsv (paste0 ("endBeds/split_" , strain, "_chrom11_toEnd_genes.tab.txt" )) %>% mutate (strain = strain ) %>% mutate (chromGlobal = gsub (paste0 (strain, "_" ), "" , col.0 )) %>% mutate (chromGlobal = gsub ("_v3" , "" , chromGlobal)) %>% rename (chrom = col.0 , start = col.1 , end = col.2 )= bind_rows (all11, strain11)# PfIT_00_10 = readr::read_tsv(paste0("possibleFragmentsOf11/split_PfIT_00_10_genes.tab.txt")) %>% # mutate(strain = "PfIT" ) %>% # mutate(chromGlobal = gsub(paste0("PfIT", "_"), "", col.0)) %>% # mutate(chromGlobal = gsub("_v3", "", chromGlobal)) %>% # rename(chrom = col.0, # start = col.1, # end = col.2) # all11 = bind_rows(all11, PfIT_00_10) # PfKH01_00_27 = readr::read_tsv(paste0("possibleFragmentsOf11/split_PfKH01_00_27_genes.tab.txt")) %>% # mutate(strain = "PfKH01" ) %>% # mutate(chromGlobal = gsub(paste0("PfKH01", "_"), "", col.0)) %>% # mutate(chromGlobal = gsub("_v3", "", chromGlobal)) %>% # rename(chrom = col.0, # start = col.1, # end = col.2) %>% # mutate(start = abs(start - 160672))%>% # mutate(end = abs(end - 160672)) # # all11 = bind_rows(all11, PfKH01_00_27) = all11 %>% filter (col.3 %!in% c ("Pf3D7_11_v3_1916391_1918050_PF3D7_1148100" , "Pf3D7_11_v3_1920267_1920811_PF3D7_1148200" , "Pf3D7_11_v3_1922655_1922928_PF3D7_1148500" )) %>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse ("membrane associated erythrocyte binding-like protein" == description, "merozoite adhesive erythrocytic binding protein" , description)) %>% mutate (description = ifelse ("membrane associated erythrocyte binding-likeprotein" == description, "merozoite adhesive erythrocytic binding protein" , description)) %>% mutate (description = ifelse ("membrane associated histidine-rich protein 1" == description, "membrane associated histidine-rich protein" , description)) %>% mutate (description = gsub (",putative" , ", putative" , description)) %>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description))%>% mutate (description = gsub ("unknownfunction" , "unknown function" , description))%>% mutate (description = gsub ("conserved protein, unknown function" , "conserved Plasmodium protein, unknown function" , description)) %>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse ("membrane associated histidine-rich protein 1" == description, "membrane associated histidine-rich protein" , description))%>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description))%>% mutate (description = gsub ("unknownfunction" , "unknown function" , description))%>% mutate (description = gsub (" \\ (SURFIN" , " \\ (SURFIN" , description)) %>% mutate (description = ifelse (grepl ("Plasmodium exported protein" , description), "Plasmodium exported protein (PHIST)" , description)) %>% mutate (description = ifelse ("erythrocyte membrane protein 1 (PfEMP1), exon 2" == description, "erythrocyte membrane protein 1 (PfEMP1), exon 2, pseudogene" , description)) %>% mutate (description = ifelse (grepl ("sporozoite and liver stage tryptophan-rich protein, putative" , description), "tryptophan/threonine-rich antigen" , description))%>% mutate (description = ifelse (grepl ("CRA domain-containing protein, putative" , description), "conserved Plasmodium protein, unknown function" , description))%>% mutate (description = gsub (",pseudogene" , ", pseudogene" , description)) %>% mutate (description = gsub ("surfaceantigen" , "surface antigen" , description)) %>% mutate (description = gsub ("Tetratricopeptide repeat, putative" , "tetratricopeptide repeat protein, putative" , description)) %>% mutate (description = gsub ("transmembraneprotein" , "transmembrane protein" , description)) %>% mutate (description = ifelse (grepl ("PfEMP1" , description) & grepl ("pseudogene" , description), "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description)) %>% mutate (description = gsub ("PIR protein" , "stevor" , description)) %>% mutate (description = gsub ("erythrocyte membrane protein 1-like" , "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description)) %>% mutate (description = gsub ("acidic terminal segments, variant surface antigen of PfEMP1, putative" , "erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description))%>% mutate (description = ifelse (grepl ("CoA binding protein" , description, ignore.case = T), "acyl-CoA binding protein" , description)) %>% mutate (description = ifelse (grepl ("transfer RNA" , description) | grepl ("tRNA" , description), "tRNA" , description))%>% mutate (description = ifelse (grepl ("cytoadherence" , description), "CLAG" , description))%>% mutate (description = ifelse (grepl ("surface-associated interspersed protein" , description), "SURFIN" , description))%>% mutate (description = ifelse (grepl ("SURFIN" , description), "SURFIN" , description))%>% mutate (description = ifelse (grepl ("stevor-like" , description), "stevor, pseudogene" , description)) %>% mutate (description = ifelse (grepl ("exported protein family" , description), "exported protein family" , description)) %>% mutate (description = ifelse (grepl ("ribosomal RNA" , description), "rRNA" , description)) %>% mutate (description = ifelse (grepl ("serine/threonine protein kinase" , description), "serine/threonine protein kinase, FIKK family" , description)) %>% mutate (description = ifelse (grepl ("hypothetical protein" , description), "hypothetical protein, conserved" , description)) %>% mutate (description = ifelse (grepl ("conserved Plasmodium protein, unknown function" , description), "hypothetical protein, conserved" , description)) %>% mutate (description = ifelse (grepl ("Rifin/stevor family, putative" , description), "stevor" , description)) %>% mutate (description = ifelse (grepl ("stevor" , description), "stevor" , description)) %>% mutate (description = ifelse (grepl ("rifin" , description), "rifin" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte membrane protein 1 (PfEMP1), pseudogene" , description), "erythrocyte membrane protein 1 (PfEMP1)" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte membrane protein 1" , description), "erythrocyte membrane protein 1 (PfEMP1)" , description)) %>% mutate (description = ifelse (grepl ("probably protein" , description), "unspecified product" , description)) %>% mutate (description = ifelse (grepl ("RESA" , description), "RESA" , description)) %>% mutate (description = ifelse (grepl ("ring-infected erythrocyte surface antigen" , description), "ring-infected erythrocyte surface antigen" , description))%>% mutate (description = ifelse (grepl ("Duffy binding domain/Erythrocyte binding antigen175, putative" , description), "erythrocyte binding like protein 1" , description)) %>% mutate (description = ifelse (grepl ("erythrocyte binding like protein 1" , description), "erythrocyte binding like protein 1" , description))%>% mutate (description = ifelse (grepl ("unspecified product" , description), "hypothetical protein, conserved" , description))%>% mutate (description = ifelse (grepl ("probable protein, unknown function" , description), "hypothetical protein, conserved" , description)) %>% mutate (description = ifelse (grepl ("RESA" , description), "ring-infected erythrocyte surface antigen" , description)) %>% mutate (description = gsub (", putative" , "" , description))= all11 %>% group_by (strain, chrom) %>% summarise (minStart = min (start))= all11 %>% left_join (all11_strainStarts) %>% mutate (globalStart = start - minStart, globalEnd = end - minStart)= chromLengths %>% filter (chrom %in% all11_strainStarts$ chrom) %>% left_join (all11_strainStarts) %>% mutate (globalStart = 0 , globalEnd = length - minStart) %>% mutate (chrom = factor (chrom, levels = chrom))= all11 %>% mutate (chrom = factor (chrom, levels = chromLengths_11$ chrom)) # %>% # mutate(globalStart = ifelse("PfKH01_00_27" == chrom, globalStart + 17822, globalStart))%>% # mutate(globalEnd = ifelse("PfKH01_00_27" == chrom, globalEnd + 17822, globalEnd)) # %>% # mutate(globalStart = ifelse("PfIT_00_10" == chrom, globalStart + 28623, globalStart))%>% # mutate(globalEnd = ifelse("PfIT_00_10" == chrom, globalEnd + 28623 , globalEnd)) # chromLengths_11 = chromLengths_11 %>% # mutate(globalStart = ifelse("PfKH01_00_27" == chrom, globalStart + 17822, globalStart))%>% # mutate(globalEnd = ifelse("PfKH01_00_27" == chrom, globalEnd + 17822, globalEnd)) # %>% # mutate(globalStart = ifelse("PfIT_00_10" == chrom, globalStart + 28623, globalStart))%>% # mutate(globalEnd = ifelse("PfIT_00_10" == chrom, globalEnd + 28623, globalEnd)) ``` # Plotting ```{r} = unique (c (all05$ description, all07$ description,all08$ description, all13$ description, all11$ description))= scheme$ hex (length (descriptionColorsNames))names (descriptionColors) = descriptionColorsNames``` ## 05 ```{r} = ggplot () + geom_rect (data = chromLengths_05, aes (xmin = globalStart, xmax = globalEnd, ymin = as.numeric (chrom) - 0.4 ,ymax = as.numeric (chrom) + 0.4 ), fill= "grey80" ) + geom_rect (data = all05, aes (xmin = globalStart, xmax = globalEnd, ymin = as.numeric (chrom) - 0.4 ,ymax = as.numeric (chrom) + 0.4 , fill = description, ID = ID, strain = strain), color = "black" ) + scale_y_continuous (breaks = 1 : max (as.numeric (chromLengths_05$ chrom)), labels = as.character (chromLengths_05$ chrom)) + scale_fill_manual ("Genes \n Description" ,values = descriptionColors[names (descriptionColors) %in% all05$ description], guide = guide_legend (nrow = 26 )) + + theme (legend.text = element_text (size = 30 ), axis.text.x = element_text (size = 30 , color = "black" ), axis.text.y = element_text (size = 30 , color = "black" ), legend.title = element_text (size = 30 , face = "bold" ), legend.box= "vertical" , legend.margin= margin (),legend.background = element_blank (),legend.box.background = element_rect (colour = "black" )) ``` ```{r} pdf ("P_Laverania_chrom05_plot.pdf" , height = 20 , width = 30 , useDingbats = F)print (chrom05_plot)dev.off ()``` ```{r} #| column: screen-inset #| fig-height: 17 #| fig-width: 27 :: ggplotly (chrom05_plot)``` ## 07 ```{r} = ggplot () + geom_rect (data = chromLengths_07, aes (xmin = globalStart, xmax = globalEnd, ymin = as.numeric (chrom) - 0.4 ,ymax = as.numeric (chrom) + 0.4 ), fill= "grey80" ) + geom_rect (data = all07, aes (xmin = globalStart, xmax = globalEnd, ymin = as.numeric (chrom) - 0.4 ,ymax = as.numeric (chrom) + 0.4 , fill = description, ID = ID, strain = strain), color = "black" ) + scale_y_continuous (breaks = 1 : max (as.numeric (chromLengths_07$ chrom)), labels = as.character (chromLengths_07$ chrom)) + scale_fill_manual ("Genes \n Description" ,values = descriptionColors[names (descriptionColors) %in% all07$ description], guide = guide_legend (nrow = 26 )) + + theme (legend.text = element_text (size = 30 ), axis.text.x = element_text (size = 30 , color = "black" ), axis.text.y = element_text (size = 30 , color = "black" ), legend.title = element_text (size = 30 , face = "bold" ), legend.box= "vertical" , legend.margin= margin (),legend.background = element_blank (),legend.box.background = element_rect (colour = "black" )) ``` ```{r} pdf ("P_Laverania_chrom07_plot.pdf" , height = 20 , width = 30 , useDingbats = F)print (chrom07_plot)dev.off ()``` ```{r} #| column: screen-inset #| fig-height: 17 #| fig-width: 27 :: ggplotly (chrom07_plot)``` ## 08 ```{r} = ggplot () + geom_rect (data = chromLengths_08, aes (xmin = globalStart, xmax = globalEnd, ymin = as.numeric (chrom) - 0.4 ,ymax = as.numeric (chrom) + 0.4 ), fill= "grey80" ) + geom_rect (data = all08, aes (xmin = globalStart, xmax = globalEnd, ymin = as.numeric (chrom) - 0.4 ,ymax = as.numeric (chrom) + 0.4 , fill = description, ID = ID, strain = strain), color = "black" ) + scale_y_continuous (breaks = 1 : max (as.numeric (chromLengths_08$ chrom)), labels = as.character (chromLengths_08$ chrom)) + scale_fill_manual ("Genes \n Description" ,values = descriptionColors[names (descriptionColors) %in% all08$ description], guide = guide_legend (nrow = 7 )) + + theme (legend.text = element_text (size = 30 ), , axis.text.x = element_text (size = 30 , color = "black" ), axis.text.y = element_text (size = 30 , color = "black" ), legend.title = element_text (size = 30 , face = "bold" ), legend.box= "vertical" , legend.margin= margin (),legend.background = element_blank (),legend.box.background = element_rect (colour = "black" )) ``` ```{r} pdf ("P_Laverania_chrom08_plot.pdf" , height = 17 , width = 27 , useDingbats = F)print (chrom08_plot)dev.off ()``` ```{r} #| column: screen-inset #| fig-height: 17 #| fig-width: 27 :: ggplotly (chrom08_plot)``` ## 13 ```{r} = ggplot () + geom_rect (data = chromLengths_13, aes (xmin = globalStart, xmax = globalEnd, ymin = as.numeric (chrom) - 0.4 ,ymax = as.numeric (chrom) + 0.4 ), fill= "grey80" ) + geom_rect (data = all13, aes (xmin = globalStart, xmax = globalEnd, ymin = as.numeric (chrom) - 0.4 ,ymax = as.numeric (chrom) + 0.4 , fill = description, ID = ID, strain = strain), color = "black" ) + scale_y_continuous (breaks = 1 : max (as.numeric (chromLengths_13$ chrom)), labels = as.character (chromLengths_13$ chrom)) + scale_fill_manual ("Genes \n Description" ,values = descriptionColors[names (descriptionColors) %in% all13$ description], guide = guide_legend (nrow = 7 )) + + theme (legend.text = element_text (size = 30 ), axis.text.x = element_text (size = 30 , color = "black" ), axis.text.y = element_text (size = 30 , color = "black" ), legend.title = element_text (size = 30 , face = "bold" ), legend.box= "vertical" , legend.margin= margin (),legend.background = element_blank (),legend.box.background = element_rect (colour = "black" )) ``` ```{r} pdf ("P_Laverania_chrom13_plot.pdf" , height = 17 , width = 27 , useDingbats = F)print (chrom13_plot)dev.off ()``` ```{r} #| column: screen-inset #| fig-height: 17 #| fig-width: 27 :: ggplotly (chrom13_plot)``` ## 11 ```{r} = ggplot () + geom_rect (data = chromLengths_11, aes (xmin = globalStart, xmax = globalEnd, ymin = as.numeric (chrom) - 0.4 ,ymax = as.numeric (chrom) + 0.4 ), fill= "grey80" ) + geom_rect (data = all11, aes (xmin = globalStart, xmax = globalEnd, ymin = as.numeric (chrom) - 0.4 ,ymax = as.numeric (chrom) + 0.4 , fill = description, ID = ID, strain = straincolor = "black" ) + scale_y_continuous (breaks = 1 : max (as.numeric (chromLengths_11$ chrom)), labels = as.character (chromLengths_11$ chrom)) + scale_fill_manual ("Genes \n Description" ,values = descriptionColors[names (descriptionColors) %in% all11$ description], guide = guide_legend (nrow = 8 )) + + theme (legend.text = element_text (size = 30 ), axis.text.x = element_text (size = 30 , color = "black" ), axis.text.y = element_text (size = 30 , color = "black" ), legend.title = element_text (size = 30 , face = "bold" ), legend.box= "vertical" , legend.margin= margin (),legend.background = element_blank (),legend.box.background = element_rect (colour = "black" )) ``` ```{r} pdf ("P_Laverania_chrom11_plot.pdf" , height = 17 , width = 27 , useDingbats = F)print (chrom11_plot)dev.off ()``` ```{r} #| column: screen-inset #| fig-height: 17 #| fig-width: 27 :: ggplotly (chrom11_plot)```